The SWI/SNF chromatin-remodeling subunit DPF2 facilitates NRF2-dependent antiinflammatory and antioxidant gene expression
- PMID: 37200093
- PMCID: PMC10313367
- DOI: 10.1172/JCI158419
The SWI/SNF chromatin-remodeling subunit DPF2 facilitates NRF2-dependent antiinflammatory and antioxidant gene expression
Abstract
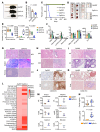
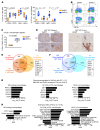
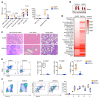
During emergency hematopoiesis, hematopoietic stem cells (HSCs) rapidly proliferate to produce myeloid and lymphoid effector cells, a response that is critical against infection or tissue injury. If unresolved, this process leads to sustained inflammation, which can cause life-threatening diseases and cancer. Here, we identify a role of double PHD fingers 2 (DPF2) in modulating inflammation. DPF2 is a defining subunit of the hematopoiesis-specific BAF (SWI/SNF) chromatin-remodeling complex, and it is mutated in multiple cancers and neurological disorders. We uncovered that hematopoiesis-specific Dpf2-KO mice developed leukopenia, severe anemia, and lethal systemic inflammation characterized by histiocytic and fibrotic tissue infiltration resembling a clinical hyperinflammatory state. Dpf2 loss impaired the polarization of macrophages responsible for tissue repair, induced the unrestrained activation of Th cells, and generated an emergency-like state of HSC hyperproliferation and myeloid cell-biased differentiation. Mechanistically, Dpf2 deficiency resulted in the loss of the BAF catalytic subunit BRG1 from nuclear factor erythroid 2-like 2-controlled (NRF2-controlled) enhancers, impairing the antioxidant and antiinflammatory transcriptional response needed to modulate inflammation. Finally, pharmacological reactivation of NRF2 suppressed the inflammation-mediated phenotypes and lethality of Dpf2Δ/Δ mice. Our work establishes an essential role of the DPF2-BAF complex in licensing NRF2-dependent gene expression in HSCs and immune effector cells to prevent chronic inflammation.
Keywords: Epigenetics; Hematology; Hematopoietic stem cells; Inflammation; Macrophages.
Figures





References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

