A nonstructural protein 1 capture enzyme-linked immunosorbent assay specific for dengue viruses
- PMID: 37200264
- PMCID: PMC10194908
- DOI: 10.1371/journal.pone.0285878
A nonstructural protein 1 capture enzyme-linked immunosorbent assay specific for dengue viruses
Abstract
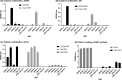
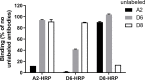
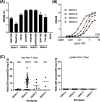
Dengue non-structural protein (NS1) is an important diagnostic marker during the acute phase of infection. Because NS1 is partially conserved across the flaviviruses, a highly specific DENV NS-1 diagnostic test is needed to differentiate dengue infection from Zika virus (ZIKV) infection. In this study, we characterized three newly isolated antibodies against NS1 (A2, D6 and D8) from a dengue-infected patient and a previously published human anti-NS1 antibody (Den3). All four antibodies recognized multimeric forms of NS1 from different serotypes. A2 bound to NS1 from DENV-1, -2, and -3, D6 bound to NS1 from DENV-1, -2, and -4, and D8 and Den3 interacted with NS1 from all four dengue serotypes. Using a competition ELISA, we found that A2 and D6 bound to overlapping epitopes on NS1 whereas D8 recognized an epitope distinct from A2 and D6. In addition, we developed a capture ELISA that specifically detected NS1 from dengue viruses, but not ZIKV, using Den3 as the capture antibody and D8 as the detecting antibody. This assay detected NS1 from all the tested dengue virus strains and dengue-infected patients. In conclusion, we established a dengue-specific capture ELISA using human antibodies against NS1. This assay has the potential to be developed as a point-of-care diagnostic tool.
Copyright: © 2023 Lim et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Organization WH. Dengue and severe dengue 2021. Available from: https://www.who.int/news-room/fact-sheets/detail/dengue-and-severe-dengue.
-
- de la Cruz-Hernandez SI, Flores-Aguilar H, Gonzalez-Mateos S, Lopez-Martinez I, Alpuche-Aranda C, Ludert JE, et al.. Determination of viremia and concentration of circulating nonstructural protein 1 in patients infected with dengue virus in Mexico. Am J Trop Med Hyg. 2013;88(3):446–54. doi: 10.4269/ajtmh.12-0023 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

