Oxalic acid blocked the binding of spike protein from SARS-CoV-2 Delta (B.1.617.2) and Omicron (B.1.1.529) variants to human angiotensin-converting enzymes 2
- PMID: 37200310
- PMCID: PMC10194883
- DOI: 10.1371/journal.pone.0285722
Oxalic acid blocked the binding of spike protein from SARS-CoV-2 Delta (B.1.617.2) and Omicron (B.1.1.529) variants to human angiotensin-converting enzymes 2
Abstract
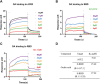
An epidemic of Corona Virus Disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is spreading worldwide. Moreover, the emergence of SARS-CoV-2 variants of concern, such as Delta and Omicron, has seriously challenged the application of current therapeutics including vaccination and drugs. Relying on interaction of spike protein with receptor angiotensin-converting enzymes 2 (ACE2), SARS-CoV-2 successfully invades to the host cells, which indicates a strategy that identification of small-molecular compounds to block the entry is of great significance for COVID-19 prevention. Our study evaluated the potential efficacy of natural compound oxalic acid (OA) as an inhibitory agent against SARS-CoV-2 invasion, particular on the interaction of the receptor binding domain (RBD) of Delta and Omicron variants to ACE2. By employing a competitive binding assay in vitro, OA significantly blocked the binding of RBDs from Delta B.1.617.2 and Omicron B.1.1.529 to ACE2, but has no effect on the wide-type SARS-CoV-2 strain. Furthermore, OA inhibited the entries of Delta and Omicron pseudovirus into ACE2 high expressing-HEK293T cells. By surface plasmon resonance (SPR) assay, the direct bindings of OA to RBD and ACE2 were analyzed and OA had both affinities with RBDs of B.1.617.2 and B.1.1.529 and with ACE2. Molecular docking predicted the binding sites on the RBD-ACE2 complex and it showed similar binding abilities to both complex of variant Delta or Omicron RBD and ACE2. In conclusion, we provided a promising novel small-molecule compound OA as an antiviral candidate by blocking the cellular entries of SARS-CoV-2 variants.
Copyright: © 2023 Wang et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures







References
-
- WHO. WHO Coronavirus (COVID-19) Dashboard. Geneva: WHO.2022. Available from: https://covid19.who.int/
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

