Brain morphometry in older adults with and without dementia using extremely rapid structural scans
- PMID: 37201641
- PMCID: PMC10330834
- DOI: 10.1016/j.neuroimage.2023.120173
Brain morphometry in older adults with and without dementia using extremely rapid structural scans
Abstract
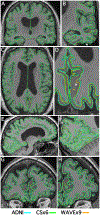
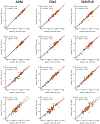
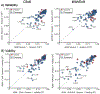
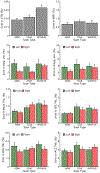
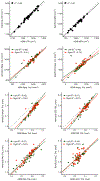
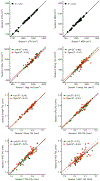
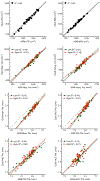
T1-weighted structural MRI is widely used to measure brain morphometry (e.g., cortical thickness and subcortical volumes). Accelerated scans as fast as one minute or less are now available but it is unclear if they are adequate for quantitative morphometry. Here we compared the measurement properties of a widely adopted 1.0 mm resolution scan from the Alzheimer's Disease Neuroimaging Initiative (ADNI = 5'12'') with two variants of highly accelerated 1.0 mm scans (compressed-sensing, CSx6 = 1'12''; and wave-controlled aliasing in parallel imaging, WAVEx9 = 1'09'') in a test-retest study of 37 older adults aged 54 to 86 (including 19 individuals diagnosed with a neurodegenerative dementia). Rapid scans produced highly reliable morphometric measures that largely matched the quality of morphometrics derived from the ADNI scan. Regions of lower reliability and relative divergence between ADNI and rapid scan alternatives tended to occur in midline regions and regions with susceptibility-induced artifacts. Critically, the rapid scans yielded morphometric measures similar to the ADNI scan in regions of high atrophy. The results converge to suggest that, for many current uses, extremely rapid scans can replace longer scans. As a final test, we explored the possibility of a 0'49'' 1.2 mm CSx6 structural scan, which also showed promise. Rapid structural scans may benefit MRI studies by shortening the scan session and reducing cost, minimizing opportunity for movement, creating room for additional scan sequences, and allowing for the repetition of structural scans to increase precision of the estimates.
Keywords: ADNI; Aging; Alzheimer's disease; Frontotemporal lobar degeneration; Hippocampus; MRI.
Copyright © 2023. Published by Elsevier Inc.
Conflict of interest statement
Declaration of Competing Interest Tom Hilbert and Tobias Kober are employed by Siemens Healthineers International AG, Switzerland. The authors have no other conflicts of interest to report.
Figures



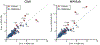


References
-
- Baum GL, Flournoy JC, Glasser MF, Harms MP, Mair P, Sanders AFP, Barch DM, Buckner RL, Bookheimer S, Dapretto M, Smith S, Thomas KM, Yacoub E, Van Essen DC, Somerville LH, 2022. Graded variation in T1w/T2w ratio during adolescence: measurement, caveats, and implications for development of cortical myelin. J. Neurosci 42, 5681–5694. doi: 10.1523/JNEUROSCI.2380-21.2022. - DOI - PMC - PubMed
-
- Bejanin A, Tammewar G, Marx G, Cobigo Y, Iaccarino L, Kornak J, Staffaroni AM, Dickerson BC, Boeve BF, Knopman DS, Gorno-Tempini M, Miller BL, Jagust WJ, Boxer AL, Rosen HJ, Rabinovici GD, 2020. Longitudinal structural and metabolic changes in frontotemporal dementia. Neurology 95, e140–e154. doi: 10.1212/WNL.0000000000009760. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

