USP28 controls SREBP2 and the mevalonate pathway to drive tumour growth in squamous cancer
- PMID: 37202505
- PMCID: PMC10307777
- DOI: 10.1038/s41418-023-01173-6
USP28 controls SREBP2 and the mevalonate pathway to drive tumour growth in squamous cancer
Abstract
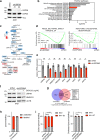
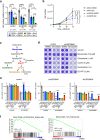
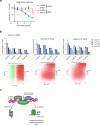
SREBP2 is a master regulator of the mevalonate pathway (MVP), a biosynthetic process that drives the synthesis of dolichol, heme A, ubiquinone and cholesterol and also provides substrates for protein prenylation. Here, we identify SREBP2 as a novel substrate for USP28, a deubiquitinating enzyme that is frequently upregulated in squamous cancers. Our results show that silencing of USP28 reduces expression of MVP enzymes and lowers metabolic flux into this pathway. We also show that USP28 binds to mature SREBP2, leading to its deubiquitination and stabilisation. USP28 depletion rendered cancer cells highly sensitive to MVP inhibition by statins, which was rescued by the addition of geranyl-geranyl pyrophosphate. Analysis of human tissue microarrays revealed elevated expression of USP28, SREBP2 and MVP enzymes in lung squamous cell carcinoma (LSCC) compared to lung adenocarcinoma (LADC). Moreover, CRISPR/Cas-mediated deletion of SREBP2 selectively attenuated tumour growth in a KRas/p53/LKB1 mutant mouse model of lung cancer. Finally, we demonstrate that statins synergise with a dual USP28/25 inhibitor to reduce viability of SCC cells. Our findings suggest that combinatorial targeting of MVP and USP28 could be a therapeutic strategy for the treatment of squamous cell carcinomas.
© 2023. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

