Premature ovarian insufficiency is associated with global alterations in the regulatory landscape and gene expression in balanced X-autosome translocations
- PMID: 37202802
- PMCID: PMC10197467
- DOI: 10.1186/s13072-023-00493-8
Premature ovarian insufficiency is associated with global alterations in the regulatory landscape and gene expression in balanced X-autosome translocations
Abstract
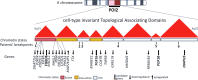
Background: Patients with balanced X-autosome translocations and premature ovarian insufficiency (POI) constitute an interesting paradigm to study the effect of chromosome repositioning. Their breakpoints are clustered within cytobands Xq13-Xq21, 80% of them in Xq21, and usually, no gene disruption can be associated with POI phenotype. As deletions within Xq21 do not cause POI, and since different breakpoints and translocations with different autosomes lead to this same gonadal phenotype, a "position effect" is hypothesized as a possible mechanism underlying POI pathogenesis.
Objective and methods: To study the effect of the balanced X-autosome translocations that result in POI, we fine-mapped the breakpoints in six patients with POI and balanced X-autosome translocations and addressed gene expression and chromatin accessibility changes in four of them.
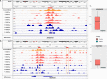
Results: We observed differential expression in 85 coding genes, associated with protein regulation, multicellular regulation, integrin signaling, and immune response pathways, and 120 differential peaks for the three interrogated histone marks, most of which were mapped in high-activity chromatin state regions. The integrative analysis between transcriptome and chromatin data pointed to 12 peaks mapped less than 2 Mb from 11 differentially expressed genes in genomic regions not related to the patients' chromosomal rearrangement, suggesting that translocations have broad effects on the chromatin structure.
Conclusion: Since a wide impact on gene regulation was observed in patients, our results observed in this study support the hypothesis of position effect as a pathogenic mechanism for premature ovarian insufficiency associated with X-autosome translocations. This work emphasizes the relevance of chromatin changes in structural variation, since it advances our knowledge of the impact of perturbations in the regulatory landscape within interphase nuclei, resulting in the position effect pathogenicity.
Keywords: Chromatin structure; Position effect; RNA sequencing; X-autosome translocation.
© 2023. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

