Low XIST expression in Sertoli cells of Klinefelter syndrome patients causes high susceptibility of these cells to an extra X chromosome
- PMID: 37202929
- PMCID: PMC10715603
- DOI: 10.4103/aja202315
Low XIST expression in Sertoli cells of Klinefelter syndrome patients causes high susceptibility of these cells to an extra X chromosome
Abstract
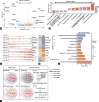
Klinefelter syndrome (KS) is the most common genetic cause of human male infertility. However, the effect of the extra X chromosome on different testicular cell types remains poorly understood. Here, we profiled testicular single-cell transcriptomes from three KS patients and normal karyotype control individuals. Among the different somatic cells, Sertoli cells showed the greatest transcriptome changes in KS patients. Further analysis showed that X-inactive-specific transcript ( XIST ), a key factor that inactivates one X chromosome in female mammals, was widely expressed in each testicular somatic cell type but not in Sertoli cells. The loss of XIST in Sertoli cells leads to an increased level of X chromosome genes, and further disrupts their transcription pattern and cellular function. This phenomenon was not detected in other somatic cells such as Leydig cells and vascular endothelial cells. These results proposed a new mechanism to explain why testicular atrophy in KS patients is heterogeneous with loss of seminiferous tubules but interstitial hyperplasia. Our study provides a theoretical basis for subsequent research and related treatment of KS by identifying Sertoli cell-specific X chromosome inactivation failure.
Copyright © 2023 Copyright: © The Author(s)(2023).
Conflict of interest statement
All authors declare no competing interests.
Supplementary Information is linked to the online version of the paper on the
Figures





References
-
- Bojesen A, Juul S, Gravholt CH. Prenatal and postnatal prevalence of Klinefelter syndrome:a national registry study. J Clin Endocrinol Metab. 2003;88:622–6. - PubMed
-
- Aksglaede L, Skakkebaek NE, Almstrup K, Juul A. Clinical and biological parameters in 166 boys, adolescents and adults with nonmosaic Klinefelter syndrome: a Copenhagen experience. Acta Paediatr. 2011;100:793–806. - PubMed
-
- Bergère M, Wainer R, Nataf V, Bailly M, Gombault M, et al. Biopsied testis cells of four 47,XXY patients: fluorescence in-situ hybridization and ICSI results. Hum Reprod. 2002;17:32–7. - PubMed
-
- Handel MA. The XY body: a specialized meiotic chromatin domain. Exp Cell Res. 2004;296:57–63. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

