This is a preprint.
Organoid modeling of lung-resident immune responses to SARS-CoV-2 infection
- PMID: 37205380
- PMCID: PMC10187413
- DOI: 10.21203/rs.3.rs-2870695/v1
Organoid modeling of lung-resident immune responses to SARS-CoV-2 infection
Abstract
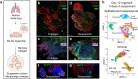
Tissue-resident immunity underlies essential host defenses against pathogens, but analysis in humans has lacked in vitro model systems where epithelial infection and accompanying resident immune cell responses can be observed en bloc. Indeed, human primary epithelial organoid cultures typically omit immune cells, and human tissue resident-memory lymphocytes are conventionally assayed without an epithelial infection component, for instance from peripheral blood, or after extraction from organs. Further, the study of resident immunity in animals can be complicated by interchange between tissue and peripheral immune compartments. To study human tissue-resident infectious immune responses in isolation from secondary lymphoid organs, we generated adult human lung three-dimensional air-liquid interface (ALI) lung organoids from intact tissue fragments that co-preserve epithelial and stromal architecture alongside endogenous lung-resident immune subsets. These included T, B, NK and myeloid cells, with CD69+CD103+ tissue-resident and CCR7- and/or CD45RA- TRM and conservation of T cell receptor repertoires, all corresponding to matched fresh tissue. SARS-CoV-2 vigorously infected organoid lung epithelium, alongside secondary induction of innate cytokine production that was inhibited by antiviral agents. Notably, SARS-CoV-2-infected organoids manifested adaptive virus-specific T cell activation that was specific for seropositive and/or previously infected donor individuals. This holistic non-reconstitutive organoid system demonstrates the sufficiency of lung to autonomously mount adaptive T cell memory responses without a peripheral lymphoid component, and represents an enabling method for the study of human tissue-resident immunity.
Conflict of interest statement
Additional Declarations: Yes there is potential Competing Interest. C.J.K. and S.S.C. are listed as inventors on provisional patent PCT/US2022/029869 describing the methods in this paper. C.J.K. is a founder of Surrozen Inc. C.J.K. and M.M.D. are founders of Mozart Therapeutics and NextVivo, Inc. All other authors declare no competing interests. COMPETING INTERESTS C.J.K. and S.S.C. are listed as inventors on provisional patent PCT/US2022/029869 describing the methods in this paper. C.J.K. is a founder of Surrozen, Inc. C.J.K. and M.M.D. are founders of Mozart Therapeutics and NextVivo, Inc. All other authors declare no competing interests.
Figures





References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

