This is a preprint.
Common Analysis of Direct RNA SequencinG CUrrently Leads to Misidentification of 5-Methylcytosine Modifications at GCU Motifs
- PMID: 37205495
- PMCID: PMC10187288
- DOI: 10.1101/2023.05.03.539298
Common Analysis of Direct RNA SequencinG CUrrently Leads to Misidentification of 5-Methylcytosine Modifications at GCU Motifs
Update in
-
Common analysis of direct RNA sequencinG CUrrently leads to misidentification of m5C at GCU motifs.Life Sci Alliance. 2023 Nov 29;7(2):e202302201. doi: 10.26508/lsa.202302201. Print 2024 Feb. Life Sci Alliance. 2023. PMID: 38030223 Free PMC article.
Abstract
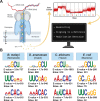
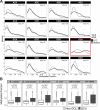
RNA modifications, such as méthylation, can be detected with Oxford Nanopore Technologies direct RNA sequencing. One commonly used tool for detecting 5-methylcytosine (m5C) modifications is Tombo, which uses an "Alternative Model" to detect putative modifications from a single sample. We examined direct RNA sequencing data from diverse taxa including virus, bacteria, fungi, and animals. The algorithm consistently identified a 5-methylcytosine at the central position of a GCU motif. However, it also identified a 5-methylcytosine in the same motif in fully unmodified in vitro transcribed RNA, suggesting that this a frequent false prediction. In the absence of further validation, several published predictions of 5-methylcytosine in human coronavirus and human cerebral organoid RNA in a GCU context should be reconsidered.
Figures
References
-
- Garalde DR, Snell EA, Jachimowicz D, Sipos B, Lloyd JH, Bruce M, Pantic N, Admassu T, James P, Warland A, Jordan M, Ciccone J, Serra S, Keenan J, Martin S, McNeill L, Wallace EJ, Jayasinghe L, Wright C, Blasco J, Young S, Brocklebank D, Juul S, Clarke J, Heron AJ, Turner DJ. 2018. Highly parallel direct RNA sequencing on an array of nanopores. Nat Methods 15:201–206. - PubMed
-
- Workman RE, Tang AD, Tang PS, Jain M, Tyson JR, Razaghi R, Zuzarte PC, Gilpatrick T, Payne A, Quick J, Sadowski N, Holmes N, de Jesus JG, Jones KL, Soulette CM, Snutch TP, Loman N, Paten B, Loose M, Simpson JT, Olsen HE, Brooks AN, Akeson M, Timp W. 2019. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat Methods 16:1297–1305. - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
