Unique Transcriptional Profiles Underlie Osteosarcomagenesis Driven by Different p53 Mutants
- PMID: 37205631
- PMCID: PMC10524763
- DOI: 10.1158/0008-5472.CAN-22-3464
Unique Transcriptional Profiles Underlie Osteosarcomagenesis Driven by Different p53 Mutants
Abstract
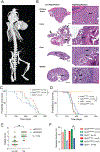
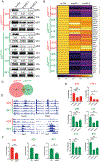
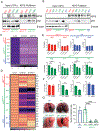
Missense mutations in the DNA binding domain of p53 are characterized as structural or contact mutations based on their effect on the conformation of the protein. These mutations show gain-of-function (GOF) activities, such as promoting increased metastatic incidence compared with p53 loss, often mediated by the interaction of mutant p53 with a set of transcription factors. These interactions are largely context specific. To understand the mechanisms by which p53 DNA binding domain mutations drive osteosarcoma progression, we created mouse models, in which either the p53 structural mutant p53R172H or the contact mutant p53R245W are expressed specifically in osteoblasts, yielding osteosarcoma tumor development. Survival significantly decreased and metastatic incidence increased in mice expressing p53 mutants compared with p53-null mice, suggesting GOF. RNA sequencing of primary osteosarcomas revealed vastly different gene expression profiles between tumors expressing the missense mutants and p53-null tumors. Further, p53R172H and p53R245W each regulated unique transcriptomes and pathways through interactions with a distinct repertoire of transcription factors. Validation assays showed that p53R245W, but not p53R172H, interacts with KLF15 to drive migration and invasion in osteosarcoma cell lines and promotes metastasis in allogeneic transplantation models. In addition, analyses of p53R248W chromatin immunoprecipitation peaks showed enrichment of KLF15 motifs in human osteoblasts. Taken together, these data identify unique mechanisms of action of the structural and contact mutants of p53.
Significance: The p53 DNA binding domain contact mutant p53R245W, but not the structural mutant p53R172H, interacts with KLF15 to drive metastasis in somatic osteosarcoma, providing a potential vulnerability in tumors expressing p53R245W mutation.
©2023 American Association for Cancer Research.
Figures



References
-
- Cho Y, Gorina S, Jeffrey PD, and Pavletich NP, Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science, 1994. 265(5170): p. 346. - PubMed
-
- Joerger AC, Allen MD, and Fersht AR, Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem, 2004. 279(2): p. 1291–6. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

