Convergent genomic signatures of local adaptation across a continental-scale environmental gradient
- PMID: 37205757
- PMCID: PMC10198635
- DOI: 10.1126/sciadv.add0560
Convergent genomic signatures of local adaptation across a continental-scale environmental gradient
Abstract
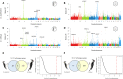
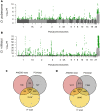
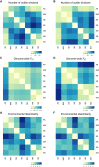
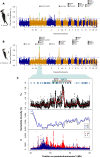
Convergent local adaptation offers a glimpse into the role of constraint and stochasticity in adaptive evolution, in particular the extent to which similar genetic mechanisms drive adaptation to common selective forces. Here, we investigated the genomics of local adaptation in two nonsister woodpeckers that are codistributed across an entire continent and exhibit remarkably convergent patterns of geographic variation. We sequenced the genomes of 140 individuals of Downy (Dryobates pubescens) and Hairy (Dryobates villosus) woodpeckers and used a suite of genomic approaches to identify loci under selection. We showed evidence that convergent genes have been targeted by selection in response to shared environmental pressures, such as temperature and precipitation. Among candidates, we found multiple genes putatively linked to key phenotypic adaptations to climate, including differences in body size (e.g., IGFPB) and plumage (e.g., MREG). These results are consistent with genetic constraints limiting the pathways of adaptation to broad climatic gradients, even after genetic backgrounds diverge.
Figures


Comment in
-
Convergent evolution in distant woodpeckers.Nat Ecol Evol. 2023 Aug;7(8):1174. doi: 10.1038/s41559-023-02109-6. Nat Ecol Evol. 2023. PMID: 37277497 No abstract available.
Similar articles
-
Multiple genomic solutions for local adaptation in two closely related species (sheep and goats) facing the same climatic constraints.Mol Ecol. 2024 Oct;33(20):e17257. doi: 10.1111/mec.17257. Epub 2023 Dec 27. Mol Ecol. 2024. PMID: 38149334
-
Landscape genomics of Quercus lobata reveals genes involved in local climate adaptation at multiple spatial scales.Mol Ecol. 2021 Jan;30(2):406-423. doi: 10.1111/mec.15731. Epub 2020 Dec 6. Mol Ecol. 2021. PMID: 33179370
-
The genomic footprint of climate adaptation in Chironomus riparius.Mol Ecol. 2018 Mar;27(6):1439-1456. doi: 10.1111/mec.14543. Epub 2018 Mar 23. Mol Ecol. 2018. PMID: 29473242
-
The Genomics of Human Local Adaptation.Trends Genet. 2020 Jun;36(6):415-428. doi: 10.1016/j.tig.2020.03.006. Epub 2020 Apr 15. Trends Genet. 2020. PMID: 32396835 Review.
-
Convergent evolution in human and domesticate adaptation to high-altitude environments.Philos Trans R Soc Lond B Biol Sci. 2019 Jul 22;374(1777):20180235. doi: 10.1098/rstb.2018.0235. Epub 2019 Jun 3. Philos Trans R Soc Lond B Biol Sci. 2019. PMID: 31154977 Free PMC article. Review.
Cited by
-
Distinct Genes with Similar Functions Underlie Convergent Evolution in Myotis Bat Ecomorphs.Mol Biol Evol. 2024 Sep 4;41(9):msae165. doi: 10.1093/molbev/msae165. Mol Biol Evol. 2024. PMID: 39116340 Free PMC article.
-
Putative climate adaptation in American pikas (Ochotona princeps) is associated with copy number variation across environmental gradients.Sci Rep. 2024 Apr 13;14(1):8568. doi: 10.1038/s41598-024-59157-6. Sci Rep. 2024. PMID: 38609461 Free PMC article.
-
Repeated global adaptation across plant species.Proc Natl Acad Sci U S A. 2024 Dec 24;121(52):e2406832121. doi: 10.1073/pnas.2406832121. Epub 2024 Dec 20. Proc Natl Acad Sci U S A. 2024. PMID: 39705310 Free PMC article.
References
-
- E. B. Rosenblum, C. E. Parent, E. E. Brandt, The molecular basis of phenotypic convergence. Annu. Rev. Ecol. Evol. Syst. 45, 203–226 (2014).
MeSH terms
LinkOut - more resources
Full Text Sources

