Cytolytic CD8+ T cells infiltrate germinal centers to limit ongoing HIV replication in spontaneous controller lymph nodes
- PMID: 37205767
- PMCID: PMC10231436
- DOI: 10.1126/sciimmunol.ade5872
Cytolytic CD8+ T cells infiltrate germinal centers to limit ongoing HIV replication in spontaneous controller lymph nodes
Abstract
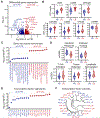
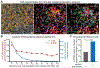
Follicular CD8+ T cells (fCD8) mediate surveillance in lymph node (LN) germinal centers against lymphotropic infections and cancers, but the precise mechanisms by which these cells mediate immune control remain incompletely resolved. To address this, we investigated functionality, clonotypic compartmentalization, spatial localization, phenotypic characteristics, and transcriptional profiles of LN-resident virus-specific CD8+ T cells in persons who control HIV without medications. Antigen-induced proliferative and cytolytic potential consistently distinguished spontaneous controllers from noncontrollers. T cell receptor analysis revealed complete clonotypic overlap between peripheral and LN-resident HIV-specific CD8+ T cells. Transcriptional analysis of LN CD8+ T cells revealed gene signatures of inflammatory chemotaxis and antigen-induced effector function. In HIV controllers, the cytotoxic effectors perforin and granzyme B were elevated among virus-specific CXCR5+ fCD8s proximate to foci of HIV RNA within germinal centers. These results provide evidence consistent with cytolytic control of lymphotropic infection supported by inflammatory recruitment, antigen-specific proliferation, and cytotoxicity of fCD8s.
Conflict of interest statement
COMPETING INTERESTS
The authors declare that they have no competing interests.
Figures





References
-
- von Andrian UH, Mempel TR, Homing and cellular traffic in lymph nodes. Nat Rev Immunol 3, 867–878 (2003). - PubMed
-
- Matloubian M, Lo CG, Cinamon G, Lesneski MJ, Xu Y, Brinkmann V, Allende ML, Proia RL, Cyster JG, Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 427, 355–360 (2004). - PubMed
-
- Shiow LR, Rosen DB, Brdickova N, Xu Y, An J, Lanier LL, Cyster JG, Matloubian M, CD69 acts downstream of interferon-alpha/beta to inhibit S1P1 and lymphocyte egress from lymphoid organs. Nature 440, 540–544 (2006). - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

