Aptamers 101: aptamer discovery and in vitro applications in biosensors and separations
- PMID: 37206388
- PMCID: PMC10189874
- DOI: 10.1039/d3sc00439b
Aptamers 101: aptamer discovery and in vitro applications in biosensors and separations
Abstract
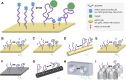
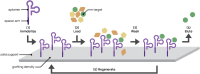
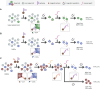
Aptamers are single-stranded nucleic acids that bind and recognize targets much like antibodies. Recently, aptamers have garnered increased interest due to their unique properties, including inexpensive production, simple chemical modification, and long-term stability. At the same time, aptamers possess similar binding affinity and specificity as their protein counterpart. In this review, we discuss the aptamer discovery process as well as aptamer applications to biosensors and separations. In the discovery section, we describe the major steps of the library selection process for aptamers, called systematic evolution of ligands by exponential enrichment (SELEX). We highlight common approaches and emerging strategies in SELEX, from starting library selection to aptamer-target binding characterization. In the applications section, we first evaluate recently developed aptamer biosensors for SARS-CoV-2 virus detection, including electrochemical aptamer-based sensors and lateral flow assays. Then we discuss aptamer-based separations for partitioning different molecules or cell types, especially for purifying T cell subsets for therapeutic applications. Overall, aptamers are promising biomolecular tools and the aptamer field is primed for expansion in biosensing and cell separation.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures







References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

