Effects of HLA single chain trimer design on peptide presentation and stability
- PMID: 37207206
- PMCID: PMC10189100
- DOI: 10.3389/fimmu.2023.1170462
Effects of HLA single chain trimer design on peptide presentation and stability
Abstract
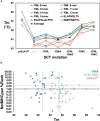
MHC class I "single-chain trimer" molecules, coupling MHC heavy chain, β2-microglobulin, and a specific peptide into a single polypeptide chain, are widely used in research. To more fully understand caveats associated with this design that may affect its use for basic and translational studies, we evaluated a set of engineered single-chain trimers with combinations of stabilizing mutations across eight different classical and non-classical human class I alleles with 44 different peptides, including a novel human/murine chimeric design. While, overall, single-chain trimers accurately recapitulate native molecules, care was needed in selecting designs for studying peptides longer or shorter than 9-mers, as single-chain trimer design could affect peptide conformation. In the process, we observed that predictions of peptide binding were often discordant with experiment and that yields and stabilities varied widely with construct design. We also developed novel reagents to improve the crystallizability of these proteins and confirmed novel modes of peptide presentation.
Keywords: HLA single-chain trimers; X-ray crystallography; human class I leukocyte antigens; pan-anti-class I antibodies; peptide presentation.
Copyright © 2023 Finton, Rupert, Friend, Dinca, Lovelace, Buerger, Rusnac, Foote-McNabb, Chour, Heath, Campbell, Pierce and Strong.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
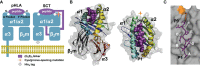
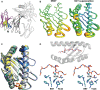


References
-
- Strong RK. Structural immunology of MHC class I proteins, homologs and receptor complexes. Modern Aspects Immunobiol (2000) 3:125–8.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

