Elevated MPP6 expression correlates with an unfavorable prognosis, angiogenesis and immune evasion in hepatocellular carcinoma
- PMID: 37207207
- PMCID: PMC10189050
- DOI: 10.3389/fimmu.2023.1173848
Elevated MPP6 expression correlates with an unfavorable prognosis, angiogenesis and immune evasion in hepatocellular carcinoma
Abstract
Background: Membrane palmitoylated proteins (MPPs) are engaged in various biological processes, such as cell adhesion and cell polarity. Dysregulated MPP members have different effects on hepatocellular carcinoma (HCC) development. However, the role of MPP6 in HCC has been unknown.
Method: HCC transcriptome and clinical data from different public databases were downloaded and analyzed, and the results were further validated by qRT-PCR, Western blotting and immunohistochemistry (IHC) using HCC cell lines and tissues. The association between MPP6 and prognosis, potential pathogenic mechanisms, angiogenesis, immune evasion, tumor mutation burden (TMB) and treatment response in HCC patients was analyzed by bioinformatics and IHC staining.
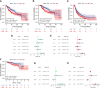
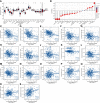
Results: MPP6 was significantly overexpressed in HCC, and its expression was related to T stage, pathologic stage, histologic grade and adverse prognosis in HCC patients. Gene set enrichment analysis revealed that differentially expressed genes were mainly enriched in the synthesis of genetic materials and the WNT signaling pathway. GEPIA database analysis and IHC staining suggested that MPP6 expression had a positive correlation with angiogenesis. Single-cell dataset analysis indicated that MPP6 was associated with features of the tumor microenvironment. Additional analyses discovered that MPP6 expression was inversely related to immune cell infiltration and was involved in tumor immune evasion. MPP6 expression was positively associated with TMB, and patients with high TMB had an adverse prognosis. Immunotherapy was more effective in HCC patients with low MPP6 expression, whereas those with high MPP6 expression responded better to sorafenib, gemcitabine, 5-FU, and doxorubicin.
Conclusions: Elevated MPP6 expression is associated with an unfavorable prognosis, angiogenesis and immune evasion in HCC. Moreover, MPP6 has the potential to be used to assess TMB and treatment response. Therefore, MPP6 might serve as a novel prognostic biomarker and therapeutic target for HCC.
Keywords: MPP6; angiogenesis; hepatocellular carcinoma; immune evasion; prognosis; treatment response.
Copyright © 2023 Cheng, Wang, Liu, Lv, Ji, Yu, Zhang and Yang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

