Dynamic accumulation of cyclobutane pyrimidine dimers and its response to changes in DNA conformation
- PMID: 37207339
- PMCID: PMC10287945
- DOI: 10.1093/nar/gkad434
Dynamic accumulation of cyclobutane pyrimidine dimers and its response to changes in DNA conformation
Abstract
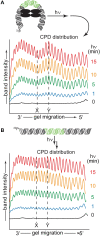
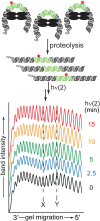
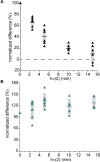
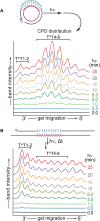
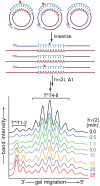
Photochemical dimerization of adjacent pyrimidines is fundamental to the creation of mutagenic hotspots caused by ultraviolet light. Distribution of the resulting lesions (cyclobutane pyrimidine dimers, CPDs) is already known to be highly variable in cells, and in vitro models have implicated DNA conformation as a major basis for this observation. Past efforts have primarily focused on mechanisms that influence CPD formation and have rarely considered contributions of CPD reversion. However, reversion is competitive under the standard conditions of 254 nm irradiation as illustrated in this report based on the dynamic response of CPDs to changes in DNA conformation. A periodic profile of CPDs was recreated in DNA held in a bent conformation by λ repressor. After linearization of this DNA, the CPD profile relaxed to its characteristic uniform distribution over a similar time of irradiation to that required to generate the initial profile. Similarly, when a T tract was released from a bent conformation, its CPD profile converted under further irradiation to that consistent with a linear T tract. This interconversion of CPDs indicates that both its formation and reversion exert control on CPD populations long before photo-steady-state conditions are achieved and suggests that the dominant sites of CPDs will evolve as DNA conformation changes in response to natural cellular processes.
© The Author(s) 2023. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
-
- Wang S.Y. Photochemistry and Photobiology of Nucleic Acids. 1976; NY: Academic Press.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

