The molecular basis of heterochromatin assembly and epigenetic inheritance
- PMID: 37207657
- PMCID: PMC10309086
- DOI: 10.1016/j.molcel.2023.04.020
The molecular basis of heterochromatin assembly and epigenetic inheritance
Abstract
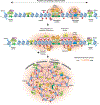
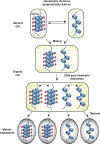
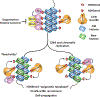
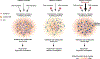
Heterochromatin plays a fundamental role in gene regulation, genome integrity, and silencing of repetitive DNA elements. Histone modifications are essential for the establishment of heterochromatin domains, which is initiated by the recruitment of histone-modifying enzymes to nucleation sites. This leads to the deposition of histone H3 lysine-9 methylation (H3K9me), which provides the foundation for building high-concentration territories of heterochromatin proteins and the spread of heterochromatin across extended domains. Moreover, heterochromatin can be epigenetically inherited during cell division in a self-templating manner. This involves a "read-write" mechanism where pre-existing modified histones, such as tri-methylated H3K9 (H3K9me3), support chromatin association of the histone methyltransferase to promote further deposition of H3K9me. Recent studies suggest that a critical density of H3K9me3 and its associated factors is necessary for the propagation of heterochromatin domains across multiple generations. In this review, I discuss the key experiments that have highlighted the importance of modified histones for epigenetic inheritance.
Keywords: epigenetic; heterochromatin; histone methylation; inheritance; read-write; silencing.
Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests The author is a member of the Molecular Cell advisory board.
Figures
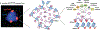
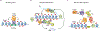

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

