A novel mutation-proof, next-generation vaccine to fight against upcoming SARS-CoV-2 variants and subvariants, designed through AI enabled approaches and tools, along with the machine learning based immune simulation: A vaccine breakthrough
- PMID: 37207746
- PMCID: PMC10188376
- DOI: 10.1016/j.ijbiomac.2023.124893
A novel mutation-proof, next-generation vaccine to fight against upcoming SARS-CoV-2 variants and subvariants, designed through AI enabled approaches and tools, along with the machine learning based immune simulation: A vaccine breakthrough
Abstract
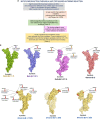
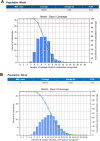
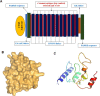
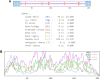
Emerging SARS-CoV-2 variants and subvariants are great concerns for their significant mutations, which are also responsible for vaccine escape. Therefore, the study was undertaken to develop a mutation-proof, next-generation vaccine to protect against all upcoming SARS-CoV-2 variants. We used advanced computational and bioinformatics approaches to develop a multi-epitopic vaccine, especially the AI model for mutation selection and machine learning (ML) strategies for immune simulation. AI enabled and the top-ranked antigenic selection approaches were used to select nine mutations from 835 RBD mutations. We selected twelve common antigenic B cell and T cell epitopes (CTL and HTL) containing the nine RBD mutations and joined them with the adjuvants, PADRE sequence, and suitable linkers. The constructs' binding affinity was confirmed through docking with TLR4/MD2 complex and showed significant binding free energy (-96.67 kcal mol-1) with positive binding affinity. Similarly, the calculated eigenvalue (2.428517e-05) from the NMA of the complex reveals proper molecular motion and superior residues' flexibility. Immune simulation shows that the candidate can induce a robust immune response. The designed mutation-proof, multi-epitopic vaccine could be a remarkable candidate for upcoming SARS-CoV-2 variants and subvariants. The study method might guide researchers in developing AI-ML and immunoinformatics-based vaccines for infectious disease.
Keywords: Immune simulation; Multi-epitopic peptide vaccine; Mutation-proof; SARS-CoV-2 variants and subvariants.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no competing interests.
Figures





References
-
- Chakraborty C., et al. SARS-CoV-2 causing pneumonia-associated respiratory disorder (COVID-19): diagnostic and proposed therapeutic options. Eur. Rev. Med. Pharmacol. Sci. 2020;24(7):4016–4026. - PubMed
-
- Chakraborty C., et al. The 2019 novel coronavirus disease (COVID-19) pandemic: a zoonotic prospective. Asian Pac. J. Trop. Med. 2020;13(6):242.
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

