ERK and USP5 govern PD-1 homeostasis via deubiquitination to modulate tumor immunotherapy
- PMID: 37208329
- PMCID: PMC10199079
- DOI: 10.1038/s41467-023-38605-3
ERK and USP5 govern PD-1 homeostasis via deubiquitination to modulate tumor immunotherapy
Erratum in
-
Author Correction: ERK and USP5 govern PD-1 homeostasis via deubiquitination to modulate tumor immunotherapy.Nat Commun. 2025 Jun 20;16(1):5357. doi: 10.1038/s41467-025-61049-w. Nat Commun. 2025. PMID: 40541949 Free PMC article. No abstract available.
Abstract
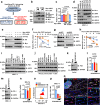
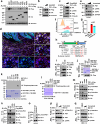
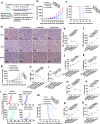
The programmed cell death protein 1 (PD-1) is an inhibitory receptor on T cells and plays an important role in promoting cancer immune evasion. While ubiquitin E3 ligases regulating PD-1 stability have been reported, deubiquitinases governing PD-1 homeostasis to modulate tumor immunotherapy remain unknown. Here, we identify the ubiquitin-specific protease 5 (USP5) as a bona fide deubiquitinase for PD-1. Mechanistically, USP5 interacts with PD-1, leading to deubiquitination and stabilization of PD-1. Moreover, extracellular signal-regulated kinase (ERK) phosphorylates PD-1 at Thr234 and promotes PD-1 interaction with USP5. Conditional knockout of Usp5 in T cells increases the production of effector cytokines and retards tumor growth in mice. USP5 inhibition in combination with Trametinib or anti-CTLA-4 has an additive effect on suppressing tumor growth in mice. Together, this study describes a molecular mechanism of ERK/USP5-mediated regulation of PD-1 and identifies potential combinatorial therapeutic strategies for enhancing anti-tumor efficacy.
© 2023. The Author(s).
Conflict of interest statement
G.J.F. has patents/pending royalties on the PD-1/PD-L1 pathway from Roche, Merck MSD, Bristol-Myers-Squibb, Merck KGA, Boehringer-Ingelheim, AstraZeneca, Dako, Leica, Mayo Clinic, Eli Lilly, and Novartis. G.J.F. has served on advisory boards for Roche, Bristol-Myers-Squibb, Xios, Origimed, Triursus, iTeos, NextPoint, IgM, Jubilant, Trillium, GV20, IOME, and Geode. G.J.F. has equity in Nextpoint, Triursus, Xios, iTeos, IgM, GV20, Invaria, and Geode. W.W. is a co-founder and consultant for ReKindle Therapeutics. The remaining authors declare no other competing interests.
Figures



References
-
- Sharma, P. & Allison, J. P. The future of immune checkpoint therapy. Science348, 56–61 (2015). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

