ADAR1-mediated RNA editing of SCD1 drives drug resistance and self-renewal in gastric cancer
- PMID: 37208334
- PMCID: PMC10199093
- DOI: 10.1038/s41467-023-38581-8
ADAR1-mediated RNA editing of SCD1 drives drug resistance and self-renewal in gastric cancer
Abstract
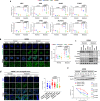
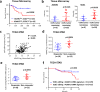
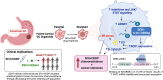
Targetable drivers governing 5-fluorouracil and cisplatin (5FU + CDDP) resistance remain elusive due to the paucity of physiologically and therapeutically relevant models. Here, we establish 5FU + CDDP resistant intestinal subtype GC patient-derived organoid lines. JAK/STAT signaling and its downstream, adenosine deaminases acting on RNA 1 (ADAR1), are shown to be concomitantly upregulated in the resistant lines. ADAR1 confers chemoresistance and self-renewal in an RNA editing-dependent manner. WES coupled with RNA-seq identify enrichment of hyper-edited lipid metabolism genes in the resistant lines. Mechanistically, ADAR1-mediated A-to-I editing on 3'UTR of stearoyl-CoA desaturase (SCD1) increases binding of KH domain-containing, RNA-binding, signal transduction-associated 1 (KHDRBS1), thereby augmenting SCD1 mRNA stability. Consequently, SCD1 facilitates lipid droplet formation to alleviate chemotherapy-induced ER stress and enhances self-renewal through increasing β-catenin expression. Pharmacological inhibition of SCD1 abrogates chemoresistance and tumor-initiating cell frequency. Clinically, high proteomic level of ADAR1 and SCD1, or high SCD1 editing/ADAR1 mRNA signature score predicts a worse prognosis. Together, we unveil a potential target to circumvent chemoresistance.
© 2023. The Author(s).
Conflict of interest statement
S.Y.L. has received research sponsorship from Pfizer, Merck, Servier, and Curegenix. The remaining authors declare no other competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

