Correcting bias in cardiac geometries derived from multimodal images using spatiotemporal mapping
- PMID: 37208380
- PMCID: PMC10199025
- DOI: 10.1038/s41598-023-33968-5
Correcting bias in cardiac geometries derived from multimodal images using spatiotemporal mapping
Erratum in
-
Publisher Correction: Correcting bias in cardiac geometries derived from multimodal images using spatiotemporal mapping.Sci Rep. 2023 Aug 21;13(1):13597. doi: 10.1038/s41598-023-40773-7. Sci Rep. 2023. PMID: 37604986 Free PMC article. No abstract available.
Abstract
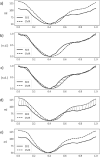
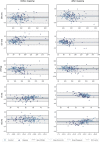
Cardiovascular imaging studies provide a multitude of structural and functional data to better understand disease mechanisms. While pooling data across studies enables more powerful and broader applications, performing quantitative comparisons across datasets with varying acquisition or analysis methods is problematic due to inherent measurement biases specific to each protocol. We show how dynamic time warping and partial least squares regression can be applied to effectively map between left ventricular geometries derived from different imaging modalities and analysis protocols to account for such differences. To demonstrate this method, paired real-time 3D echocardiography (3DE) and cardiac magnetic resonance (CMR) sequences from 138 subjects were used to construct a mapping function between the two modalities to correct for biases in left ventricular clinical cardiac indices, as well as regional shape. Leave-one-out cross-validation revealed a significant reduction in mean bias, narrower limits of agreement, and higher intraclass correlation coefficients for all functional indices between CMR and 3DE geometries after spatiotemporal mapping. Meanwhile, average root mean squared errors between surface coordinates of 3DE and CMR geometries across the cardiac cycle decreased from 7 ± 1 to 4 ± 1 mm for the total study population. Our generalised method for mapping between time-varying cardiac geometries obtained using different acquisition and analysis protocols enables the pooling of data between modalities and the potential for smaller studies to leverage large population databases for quantitative comparisons.
© 2023. The Author(s).
Conflict of interest statement
MPN is a consultant to HeartLab (NZ) Ltd. The remaining authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

