Combatting over-specialization bias in growing chemical databases
- PMID: 37208694
- PMCID: PMC10197453
- DOI: 10.1186/s13321-023-00716-w
Combatting over-specialization bias in growing chemical databases
Abstract
Background: Predicting in advance the behavior of new chemical compounds can support the design process of new products by directing the research toward the most promising candidates and ruling out others. Such predictive models can be data-driven using Machine Learning or based on researchers' experience and depend on the collection of past results. In either case: models (or researchers) can only make reliable assumptions about compounds that are similar to what they have seen before. Therefore, consequent usage of these predictive models shapes the dataset and causes a continuous specialization shrinking the applicability domain of all trained models on this dataset in the future, and increasingly harming model-based exploration of the space.
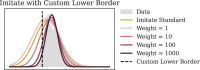
Proposed solution: In this paper, we propose CANCELS (CounterActiNg Compound spEciaLization biaS), a technique that helps to break the dataset specialization spiral. Aiming for a smooth distribution of the compounds in the dataset, we identify areas in the space that fall short and suggest additional experiments that help bridge the gap. Thereby, we generally improve the dataset quality in an entirely unsupervised manner and create awareness of potential flaws in the data. CANCELS does not aim to cover the entire compound space and hence retains a desirable degree of specialization to a specified research domain.
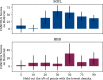
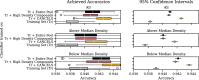
Results: An extensive set of experiments on the use-case of biodegradation pathway prediction not only reveals that the bias spiral can indeed be observed but also that CANCELS produces meaningful results. Additionally, we demonstrate that mitigating the observed bias is crucial as it cannot only intervene with the continuous specialization process, but also significantly improves a predictor's performance while reducing the number of required experiments. Overall, we believe that CANCELS can support researchers in their experimentation process to not only better understand their data and potential flaws, but also to grow the dataset in a sustainable way. All code is available under github.com/KatDost/Cancels .
Keywords: Bias; Chemical compound space; Data quality; Machine learning.
© 2023. The Author(s).
Conflict of interest statement
Jörg Wicker (co-founder, CTO) and Katharina Dost are employees of enviPath UG & Co. KG, a scientific software development company that develops and maintains the enviPath system. The authors declare no competing interests.
Figures







References
LinkOut - more resources
Full Text Sources
Miscellaneous

