Site-specific R-loops induce CGG repeat contraction and fragile X gene reactivation
- PMID: 37209683
- PMCID: PMC11505655
- DOI: 10.1016/j.cell.2023.04.035
Site-specific R-loops induce CGG repeat contraction and fragile X gene reactivation
Abstract
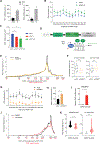
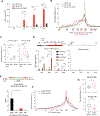
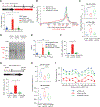
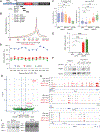
Here, we describe an approach to correct the genetic defect in fragile X syndrome (FXS) via recruitment of endogenous repair mechanisms. A leading cause of autism spectrum disorders, FXS results from epigenetic silencing of FMR1 due to a congenital trinucleotide (CGG) repeat expansion. By investigating conditions favorable to FMR1 reactivation, we find MEK and BRAF inhibitors that induce a strong repeat contraction and full FMR1 reactivation in cellular models. We trace the mechanism to DNA demethylation and site-specific R-loops, which are necessary and sufficient for repeat contraction. A positive feedback cycle comprising demethylation, de novo FMR1 transcription, and R-loop formation results in the recruitment of endogenous DNA repair mechanisms that then drive excision of the long CGG repeat. Repeat contraction is specific to FMR1 and restores the production of FMRP protein. Our study therefore identifies a potential method of treating FXS in the future.
Keywords: CGG repeat; DNA methylation; DNA repair; FMR1; FMRP; R-loop; X-linked disease; autism spectrum disorders; fragile X syndrome; gene editing; gene reactivation; neurodevelopmental disorders; repeat contraction; repeat expansion disorders; trinucleotide repeat.
Copyright © 2023 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests J.T.L. is a cofounder of Fulcrum Therapeutics and is an Advisor to Skyhawk Therapeutics. A patent application related to the technologies in this work has been filed with the USPTO.
Figures



References
-
- Berry-Kravis EM, Harnett MD, Reines SA, Reese MA, Ethridge LE, Outterson AH, Michalak C, Furman J, and Gurney ME (2021). Inhibition of phosphodiesterase-4D in adults with fragile X syndrome: a randomized, placebo-controlled, phase 2 clinical trial. Nat Med 27, 862–870. 10.1038/s41591-021-01321-w. - DOI - PubMed
-
- Berry-Kravis EM, Lindemann L, Jonch AE, Apostol G, Bear MF, Carpenter RL, Crawley JN, Curie A, Des Portes V, Hossain F, et al. (2018). Drug development for neurodevelopmental disorders: lessons learned from fragile X syndrome. Nat Rev Drug Discov 17, 280–299. 10.1038/nrd.2017.221. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

