Drug repositioning targeting glutaminase reveals drug candidates for the treatment of Alzheimer's disease patients
- PMID: 37210557
- PMCID: PMC10199278
- DOI: 10.1186/s12967-023-04192-6
Drug repositioning targeting glutaminase reveals drug candidates for the treatment of Alzheimer's disease patients
Abstract
Background: Despite numerous clinical trials and decades of endeavour, there is still no effective cure for Alzheimer's disease. Computational drug repositioning approaches may be employed for the development of new treatment strategies for Alzheimer's patients since an extensive amount of omics data has been generated during pre-clinical and clinical studies. However, targeting the most critical pathophysiological mechanisms and determining drugs with proper pharmacodynamics and good efficacy are equally crucial in drug repurposing and often imbalanced in Alzheimer's studies.
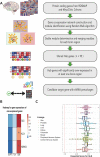
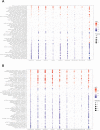
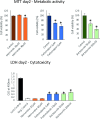
Methods: Here, we investigated central co-expressed genes upregulated in Alzheimer's disease to determine a proper therapeutic target. We backed our reasoning by checking the target gene's estimated non-essentiality for survival in multiple human tissues. We screened transcriptome profiles of various human cell lines perturbed by drug induction (for 6798 compounds) and gene knockout using data available in the Connectivity Map database. Then, we applied a profile-based drug repositioning approach to discover drugs targeting the target gene based on the correlations between these transcriptome profiles. We evaluated the bioavailability, functional enrichment profiles and drug-protein interactions of these repurposed agents and evidenced their cellular viability and efficacy in glial cell culture by experimental assays and Western blotting. Finally, we evaluated their pharmacokinetics to anticipate to which degree their efficacy can be improved.
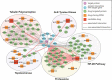
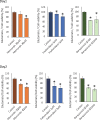
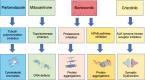
Results: We identified glutaminase as a promising drug target. Glutaminase overexpression may fuel the glutamate excitotoxicity in neurons, leading to mitochondrial dysfunction and other neurodegeneration hallmark processes. The computational drug repurposing revealed eight drugs: mitoxantrone, bortezomib, parbendazole, crizotinib, withaferin-a, SA-25547 and two unstudied compounds. We demonstrated that the proposed drugs could effectively suppress glutaminase and reduce glutamate production in the diseased brain through multiple neurodegeneration-associated mechanisms, including cytoskeleton and proteostasis. We also estimated the human blood-brain barrier permeability of parbendazole and SA-25547 using the SwissADME tool.
Conclusions: This study method effectively identified an Alzheimer's disease marker and compounds targeting the marker and interconnected biological processes by use of multiple computational approaches. Our results highlight the importance of synaptic glutamate signalling in Alzheimer's disease progression. We suggest repurposable drugs (like parbendazole) with well-evidenced activities that we linked to glutamate synthesis hereby and novel molecules (SA-25547) with estimated mechanisms for the treatment of Alzheimer's patients.
Keywords: Alzheimer’s disease; Anti-carcinogenic drugs; Gene co-expression network analysis; Glutaminase; Parbendazole; Profile-based computational drug repositioning.
© 2023. The Author(s).
Conflict of interest statement
AM and SS are co-founders of Bash Biotech Inc, USA and XL is employed by Bash Biotech Inc, USA. The other authors declare no competing interest.
Figures


Similar articles
-
Drug repositioning based on mutual information for the treatment of Alzheimer's disease patients.Med Biol Eng Comput. 2025 Aug;63(8):2249-2257. doi: 10.1007/s11517-025-03325-x. Epub 2025 Feb 17. Med Biol Eng Comput. 2025. PMID: 39961913
-
Repurposed Drugs as Potential Therapeutic Candidates for the Management of Alzheimer's Disease.Curr Drug Metab. 2017;18(9):842-852. doi: 10.2174/1389200218666170607101622. Curr Drug Metab. 2017. PMID: 28595531 Review.
-
Drug Repurposing for Alzheimer's Disease Based on Protein-Protein Interaction Network.Biomed Res Int. 2021 Oct 14;2021:1280237. doi: 10.1155/2021/1280237. eCollection 2021. Biomed Res Int. 2021. PMID: 34692825 Free PMC article.
-
Single-cell-led drug repurposing for Alzheimer's disease.Sci Rep. 2023 Jan 5;13(1):222. doi: 10.1038/s41598-023-27420-x. Sci Rep. 2023. PMID: 36604493 Free PMC article.
-
Drugs repurposing in the experimental models of Alzheimer's disease.Inflammopharmacology. 2025 Jan;33(1):195-214. doi: 10.1007/s10787-024-01608-7. Epub 2025 Jan 3. Inflammopharmacology. 2025. PMID: 39752040 Free PMC article. Review.
Cited by
-
DeepDrug as an expert guided and AI driven drug repurposing methodology for selecting the lead combination of drugs for Alzheimer's disease.Sci Rep. 2025 Jan 15;15(1):2093. doi: 10.1038/s41598-025-85947-7. Sci Rep. 2025. PMID: 39814937 Free PMC article.
-
NF-κB Pathway and Its Inhibitors: A Promising Frontier in the Management of Alzheimer's Disease.Biomedicines. 2023 Sep 21;11(9):2587. doi: 10.3390/biomedicines11092587. Biomedicines. 2023. PMID: 37761028 Free PMC article. Review.
-
Glutaminase inhibition as potential cancer therapeutics: current status and future applications.J Enzyme Inhib Med Chem. 2024 Dec;39(1):2290911. doi: 10.1080/14756366.2023.2290911. Epub 2023 Dec 11. J Enzyme Inhib Med Chem. 2024. PMID: 38078371 Free PMC article. Review.
-
Glutamate: Molecular Mechanisms and Signaling Pathway in Alzheimer's Disease, a Potential Therapeutic Target.Molecules. 2024 Dec 5;29(23):5744. doi: 10.3390/molecules29235744. Molecules. 2024. PMID: 39683904 Free PMC article. Review.
-
Drug repositioning based on mutual information for the treatment of Alzheimer's disease patients.Med Biol Eng Comput. 2025 Aug;63(8):2249-2257. doi: 10.1007/s11517-025-03325-x. Epub 2025 Feb 17. Med Biol Eng Comput. 2025. PMID: 39961913
References
-
- Ko Y. Computational drug repositioning: current progress and challenges. Appl Sci. 2020;10:5076. doi: 10.3390/app10155076. - DOI
-
- Vossel K, Ranasinghe KG, Beagle AJ, La A, Ah Pook K, Castro M, et al. Effect of levetiracetam on cognition in patients with Alzheimer disease with and without epileptiform activity: a randomized clinical trial. JAMA Neurol. 2021;78(11):1345–1354. doi: 10.1001/jamaneurol.2021.3310. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

