Non-canonical and developmental roles of the TCA cycle in plants
- PMID: 37210789
- PMCID: PMC10524895
- DOI: 10.1016/j.pbi.2023.102382
Non-canonical and developmental roles of the TCA cycle in plants
Abstract
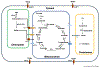
Over recent years, our understanding of the tricarboxylic acid cycle (TCAC) in living organisms has expanded beyond its canonical role in cellular energy production. In plants, TCAC metabolites and related enzymes have important roles in physiology, including vacuolar function, chelation of metals and nutrients, photorespiration, and redox regulation. Research in other organisms, including animals, has demonstrated unexpected functions of the TCAC metabolites in a number of biological processes, including signaling, epigenetic regulation, and cell differentiation. Here, we review the recent progress in discovery of non-canonical roles of the TCAC. We then discuss research on these metabolites in the context of plant development, with a focus on research related to tissue-specific functions of the TCAC. Additionally, we review research describing connections between TCAC metabolites and phytohormone signaling pathways. Overall, we discuss the opportunities and challenges in discovering new functions of TCAC metabolites in plants.
Keywords: Phytohormones; Plant development; Primary metabolism; Tricarboxylic acid cycle.
Copyright © 2023. Published by Elsevier Ltd.
Conflict of interest statement
Declaration of competing interest The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Alexandra J. Dickinson reports financial support was provided by National Institutes of Health. Alexandra J. Dickinson reports financial support was provided by National Science Foundation. Alexandra J. Dickinson reports financial support was provided by Hellman Foundation.
Figures

References
-
- Andreo CS, Gonzalez DH, Iglesias AA: Higher plant phosphoenolpyruvate carboxylase: structure and regulation. FEBS (Fed Eur Biochem Soc) Lett 1987, 213:1–8.
-
- Chollet R, Vidal J, O’Leary MH: Phosphoenolpyruvate carboxylase: a ubiquitous, highly regulated enzyme in plants. Annu Rev Plant Physiol Plant Mol Biol 1996, 47:273–298. - PubMed
-
- Lepiniec L, Thomas M, Vidal J: From enzyme activity to plant biotechnology: 30 years of research on phosphoenolpyruvate carboxylase. Plant Physiol Biochem 2003, 41:533–539.
-
- Izui K, Matsumura H, Furumoto T, Kai Y: Phosphoenolpyruvate carboxylase: a new era of structural biology. Annu Rev Plant Biol 2004, 55:69–84. - PubMed

