Clinical utility of SARS-CoV-2 subgenomic RT-PCR in a pediatric quaternary care setting
- PMID: 37210881
- PMCID: PMC10191725
- DOI: 10.1016/j.jcv.2023.105494
Clinical utility of SARS-CoV-2 subgenomic RT-PCR in a pediatric quaternary care setting
Abstract
Background: During active transcription, SARS-CoV-2 generates subgenomic regions of viral RNA. While standard SARS-CoV-2 RT-PCR amplifies region(s) of genomic RNA, it cannot distinguish active infection from remnant viral genomic material. However, screening for subgenomic RNA (sgRNA) by RT-PCR may aid in the determination of actively transcribing virus.
Objectives: To evaluate the clinical utility of SARS-CoV-2 sgRNA RT-PCR testing in a pediatric population.
Study design: Retrospective analysis was performed on inpatients from February-September 2022 positive for SARS-CoV-2 by RT-PCR with a concomitant order for sgRNA RT-PCR. Chart abstractions were conducted to determine clinical outcomes, management, and infection prevention and control (IPC) practices.
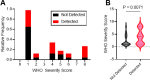
Results: Of 95 SARS-CoV-2 positive samples from 75 unique patients, 27 (28.4%) were positive by sgRNA RT-PCR. A negative sgRNA RT-PCR test allowed for de-isolation in 68 (71.6%) patient episodes. Regardless of age or sex, a positive sgRNA RT-PCR result significantly correlated with disease severity (P = 0.007), generalized COVID-19 symptoms (P = 0.012), hospitalization for COVID-19 (P = 0.019), and immune status (P = 0.024). Moreover, sgRNA RT-PCR results prompted changes in management in 28 patients (37.3%); specifically, therapeutic escalation in 13/27 (48.1%) positives and de-escalation in 15/68 (22.1%) negatives.
Conclusions: Taken together, these findings underscore the clinical utility of sgRNA RT-PCR testing in a pediatric population as we report significant associations between sgRNA RT-PCR results and clinical parameters related to COVID-19. These findings align with the proposed use of sgRNA RT-PCR testing to guide patient management and IPC practices in the hospital setting.
Keywords: Clinical utility; PCR; SARS-CoV-2; Subgenomic.
Copyright © 2023 Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Phuphuakrat A., Pasomsub E., Srichatrapimuk S., Kirdlarp S., Suksatu A., Srisaowakarn C., Manopwisedjaroen S., Ludowyke N., Purwono P.B., Priengprom T., Wongsa A., Thakkinstian A., Hongeng S., Malathum K., Thitithanyanont A., Tassaneetrithep B. Detectable Duration of Viable SARS-CoV-2, Total and Subgenomic SARS-CoV-2 RNA in Noncritically Ill COVID-19 Patients: a Prospective Cohort Study. Microbiol Spectr. 2022;10:e00503–e00522. doi: 10.1128/spectrum.00503-22. - DOI - PMC - PubMed
-
- Truong T.T., Ryutov A., Pandey U., Yee R., Goldberg L., Bhojwani D., Aguayo-Hiraldo P., Pinsky B.A., Pekosz A., Shen L., Boyd S.D., Wirz O.F., Röltgen K., Bootwalla M., Maglinte D.T., Ostrow D., Ruble D., Han J.H., Biegel J.A., Li M., Huang C., Sahoo M.K., Pannaraj P.S., O'Gorman M., Judkins A.R., Gai X., Dien Bard J. Increased viral variants in children and young adults with impaired humoral immunity and persistent SARS-CoV-2 infection: a consecutive case series. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103355. - DOI - PMC - PubMed
-
- Kim J.Y., Bae J.-.Y., Bae S., Cha H.H., Kwon J.-.S., Suh M.H., Lee H.J., Jung J., Kim M.J., Cui C., Park H., Lee J., Park M.-.S., Kim S.-.H. Diagnostic usefulness of subgenomic RNA detection of viable SARS-CoV-2 in patients with COVID-19. Clin Microbiol Infect. 2022;28:101–106. doi: 10.1016/j.cmi.2021.08.009. - DOI - PMC - PubMed
-
- Perera R.A.P.M., Tso E., Tsang O.T.Y., Tsang D.N.C., Fung K., Leung Y.W.Y., Chin A.W.H., Chu D.K.W., Cheng S.M.S., Poon L.L.M., Chuang V.W.M., Peiris M. SARS-CoV-2 Virus Culture and Subgenomic RNA for Respiratory Specimens from Patients with Mild Coronavirus Disease. Emerg Infect Dis. 2020;26:2701–2704. doi: 10.3201/eid2611.203219. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

