A New Cellular Interactome of SARS-CoV-2 Nucleocapsid Protein and Its Biological Implications
- PMID: 37211047
- PMCID: PMC10198743
- DOI: 10.1016/j.mcpro.2023.100579
A New Cellular Interactome of SARS-CoV-2 Nucleocapsid Protein and Its Biological Implications
Abstract
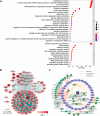
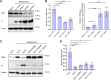
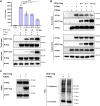
There is still much to uncover regarding the molecular details of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. As the most abundant protein, coronavirus nucleocapsid (N) protein encapsidates viral RNAs, serving as the structural component of ribonucleoprotein and virion, and participates in transcription, replication, and host regulations. Virus-host interaction might give clues to better understand how the virus affects or is affected by its host during infection and identify promising therapeutic candidates. Considering the critical roles of N, we here established a new cellular interactome of SARS-CoV-2 N by using a high-specific affinity purification (S-pulldown) assay coupled with quantitative mass spectrometry and immunoblotting validations, uncovering many N-interacting host proteins unreported previously. Bioinformatics analysis revealed that these host factors are mainly involved in translation regulations, viral transcription, RNA processes, stress responses, protein folding and modification, and inflammatory/immune signaling pathways, in line with the supposed actions of N in viral infection. Existing pharmacological cellular targets and the directing drugs were then mined, generating a drug-host protein network. Accordingly, we experimentally identified several small-molecule compounds as novel inhibitors against SARS-CoV-2 replication. Furthermore, a newly identified host factor, DDX1, was verified to interact and colocalize with N mainly by binding to the N-terminal domain of the viral protein. Importantly, loss/gain/reconstitution-of-function experiments showed that DDX1 acts as a potent anti-SARS-CoV-2 host factor, inhibiting the viral replication and protein expression. The N-targeting and anti-SARS-CoV-2 abilities of DDX1 are consistently independent of its ATPase/helicase activity. Further mechanism studies revealed that DDX1 impedes multiple activities of N, including the N-N interaction, N oligomerization, and N-viral RNA binding, thus likely inhibiting viral propagation. These data provide new clues to better depiction of the N-cell interactions and SARS-CoV-2 infection and may help inform the development of new therapeutic candidates.
Keywords: DDX1; SARS-COV-2; antiviral drug; host restriction factor; interactome; mass spectrometry; nucleocapsid (N) protein; virus–host interaction.
Copyright © 2023 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest The authors declare no competing interests.
Figures




Similar articles
-
Arginine methylation of SARS-Cov-2 nucleocapsid protein regulates RNA binding, its ability to suppress stress granule formation, and viral replication.J Biol Chem. 2021 Jul;297(1):100821. doi: 10.1016/j.jbc.2021.100821. Epub 2021 May 23. J Biol Chem. 2021. PMID: 34029587 Free PMC article.
-
MOV10 Helicase Interacts with Coronavirus Nucleocapsid Protein and Has Antiviral Activity.mBio. 2021 Oct 26;12(5):e0131621. doi: 10.1128/mBio.01316-21. Epub 2021 Sep 14. mBio. 2021. PMID: 34517762 Free PMC article.
-
Host Cellular RNA Helicases Regulate SARS-CoV-2 Infection.J Virol. 2022 Mar 23;96(6):e0000222. doi: 10.1128/jvi.00002-22. Epub 2022 Feb 2. J Virol. 2022. PMID: 35107372 Free PMC article.
-
Properties of Coronavirus and SARS-CoV-2.Malays J Pathol. 2020 Apr;42(1):3-11. Malays J Pathol. 2020. PMID: 32342926 Review.
-
Protein post-translational modification in SARS-CoV-2 and host interaction.Front Immunol. 2023 Jan 13;13:1068449. doi: 10.3389/fimmu.2022.1068449. eCollection 2022. Front Immunol. 2023. PMID: 36713387 Free PMC article. Review.
Cited by
-
Dynamic Cellular Proteome Remodeling during SARS-CoV-2 Infection. Identification of Plasma Protein Readouts.J Proteome Res. 2025 Jan 3;24(1):171-188. doi: 10.1021/acs.jproteome.4c00566. Epub 2024 Nov 26. J Proteome Res. 2025. PMID: 39593238 Free PMC article.
-
Human Helicase DDX5 is Hijacked by SARS-CoV‑2 Nsp13 Helicase to Enhance RNA Unwinding.ACS Omega. 2025 Jul 31;10(31):34941-34950. doi: 10.1021/acsomega.5c04271. eCollection 2025 Aug 12. ACS Omega. 2025. PMID: 40821550 Free PMC article.
-
Interactome profiling of Crimean-Congo hemorrhagic fever virus glycoproteins.Nat Commun. 2023 Nov 14;14(1):7365. doi: 10.1038/s41467-023-43206-1. Nat Commun. 2023. PMID: 37963884 Free PMC article.
-
Coronavirus nucleocapsid protein enhances the binding of p-PKCα to RACK1: Implications for inhibition of nucleocytoplasmic trafficking and suppression of the innate immune response.PLoS Pathog. 2024 Nov 27;20(11):e1012097. doi: 10.1371/journal.ppat.1012097. eCollection 2024 Nov. PLoS Pathog. 2024. PMID: 39602452 Free PMC article.
-
Host protein ARF1 is a proviral factor for SARS-CoV-2 and a candidate broad-spectrum therapeutic target.Nat Commun. 2025 Jul 9;16(1):6326. doi: 10.1038/s41467-025-61431-8. Nat Commun. 2025. PMID: 40634337 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

