Development of PREDAC-H1pdm to model the antigenic evolution of influenza A/(H1N1) pdm09 viruses
- PMID: 37211247
- PMCID: PMC10436056
- DOI: 10.1016/j.virs.2023.05.008
Development of PREDAC-H1pdm to model the antigenic evolution of influenza A/(H1N1) pdm09 viruses
Abstract
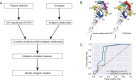
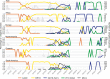
The Influenza A (H1N1) pdm09 virus caused a global pandemic in 2009 and has circulated seasonally ever since. As the continual genetic evolution of hemagglutinin in this virus leads to antigenic drift, rapid identification of antigenic variants and characterization of the antigenic evolution are needed. In this study, we developed PREDAC-H1pdm, a model to predict antigenic relationships between H1N1pdm viruses and identify antigenic clusters for post-2009 pandemic H1N1 strains. Our model performed well in predicting antigenic variants, which was helpful in influenza surveillance. By mapping the antigenic clusters for H1N1pdm, we found that substitutions on the Sa epitope were common for H1N1pdm, whereas for the former seasonal H1N1, substitutions on the Sb epitope were more common in antigenic evolution. Additionally, the localized epidemic pattern of H1N1pdm was more obvious than that of the former seasonal H1N1, which could make vaccine recommendation more sophisticated. Overall, the antigenic relationship prediction model we developed provides a rapid determination method for identifying antigenic variants, and the further analysis of evolutionary and epidemic characteristics can facilitate vaccine recommendations and influenza surveillance for H1N1pdm.
Keywords: Antigenic evolution; Antigenic relationship prediction; H1N1pdm virus; Vaccine recommendation.
Copyright © 2023 The Authors. Publishing services by Elsevier B.V. All rights reserved.
Figures


Similar articles
-
Evolution of the hemagglutinin expressed by human influenza A(H1N1)pdm09 and A(H3N2) viruses circulating between 2008-2009 and 2013-2014 in Germany.Int J Med Microbiol. 2015 Oct;305(7):762-75. doi: 10.1016/j.ijmm.2015.08.030. Epub 2015 Aug 21. Int J Med Microbiol. 2015. PMID: 26416089
-
Assessing antigenic drift and phylogeny of influenza A (H1N1) pdm09 virus in Kenya using HA1 sub-unit of the hemagglutinin gene.PLoS One. 2020 Feb 11;15(2):e0228029. doi: 10.1371/journal.pone.0228029. eCollection 2020. PLoS One. 2020. PMID: 32045419 Free PMC article.
-
Identification of amino acid substitutions supporting antigenic change of influenza A(H1N1)pdm09 viruses.J Virol. 2015 Apr;89(7):3763-75. doi: 10.1128/JVI.02962-14. Epub 2015 Jan 21. J Virol. 2015. PMID: 25609810 Free PMC article.
-
Genetic and antigenic characterization of influenza A(H1N1)pdm09 in Yantai, China, during the 2009-2017 influenza season.J Med Virol. 2019 Mar;91(3):351-360. doi: 10.1002/jmv.25328. Epub 2018 Oct 15. J Med Virol. 2019. PMID: 30267587
-
Antigenic characterization of influenza and SARS-CoV-2 viruses.Anal Bioanal Chem. 2022 Apr;414(9):2841-2881. doi: 10.1007/s00216-021-03806-6. Epub 2021 Dec 14. Anal Bioanal Chem. 2022. PMID: 34905077 Free PMC article. Review.
Cited by
-
Genetic characteristics of H1N1 influenza virus outbreak in China in early 2023.Virol Sin. 2024 Jun;39(3):520-523. doi: 10.1016/j.virs.2024.05.003. Epub 2024 May 18. Virol Sin. 2024. PMID: 38768710 Free PMC article.
References
-
- Bouckaert R., Vaughan T.G., Barido-Sottani J., Duchêne S., Fourment M., Gavryushkina A., Heled J., Jones G., Kühnert D., De Maio N., Matschiner M., Mendes F.K., Müller N.F., Ogilvie H.A., Du Plessis L., Popinga A., Rambaut A., Rasmussen D., Siveroni I., Suchard M.A., Wu C.H., Xie D., Zhang C., Stadler T., Drummond A.J. Beast 2.5: an advanced software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 2019;15:1–28. - PMC - PubMed
-
- Caton A.J., Brownlee G.G., Yewdell J.W., Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

