YBX1-Mediated DNA Methylation-Dependent SHANK3 Expression in PBMCs and Developing Cortical Interneurons in Schizophrenia
- PMID: 37211699
- PMCID: PMC10369273
- DOI: 10.1002/advs.202300455
YBX1-Mediated DNA Methylation-Dependent SHANK3 Expression in PBMCs and Developing Cortical Interneurons in Schizophrenia
Abstract
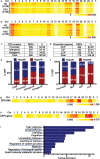
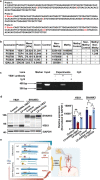
Schizophrenia (SCZ) is a severe psychiatric and neurodevelopmental disorder. The pathological process of SCZ starts early during development, way before the first onset of psychotic symptoms. DNA methylation plays an important role in regulating gene expression and dysregulated DNA methylation is involved in the pathogenesis of various diseases. The methylated DNA immunoprecipitation-chip (MeDIP-chip) is performed to investigate genome-wide DNA methylation dysregulation in peripheral blood mononuclear cells (PBMCs) of patients with first-episode SCZ (FES). Results show that the SHANK3 promoter is hypermethylated, and this hypermethylation (HyperM) is negatively correlated with the cortical surface area in the left inferior temporal cortex and positively correlated with the negative symptom subscores in FES. The transcription factor YBX1 is further found to bind to the HyperM region of SHANK3 promoter in induced pluripotent stem cells (iPSCs)-derived cortical interneurons (cINs) but not glutamatergic neurons. Furthermore, a direct and positive regulatory effect of YBX1 on the expression of SHANK3 is confirmed in cINs using shRNAs. In summary, the dysregulated SHANK3 expression in cINs suggests the potential role of DNA methylation in the neuropathological mechanism underlying SCZ. The results also suggest that HyperM of SHANK3 in PBMCs can serve as a potential peripheral biomarker of SCZ.
Keywords: DNA methylation; SHANK3; YBX1; cortical interneurons; induced pluripotent stem cells; schizophrenia.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- American Psychiatric Association ,Diagnostic and statistical manual of mental disorders: DSM‐5TM (5th ed.), American Psychiatric Publishing, Washington, DC 2013.
-
- Pepper E., Cardno G. A., Curr. Psychiatry Rev. 2014, 10, 133.
-
- Schizophrenia Working Group of the Psychiatric Genomics Consortium , Nature 2014, 511, 421.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
