LncRNA BCLET variant confers bladder cancer susceptibility through alternative splicing of MSANTD2 exon 1
- PMID: 37211917
- PMCID: PMC10358201
- DOI: 10.1002/cam4.6072
LncRNA BCLET variant confers bladder cancer susceptibility through alternative splicing of MSANTD2 exon 1
Abstract
Background: Alternative splicing (AS)-related single nucleotide polymorphisms (SNPs) are associated with risk of cancers, but the potential mechanism has not been fully elucidated.
Methods: Two-stage case-control studies comprising 1630 cases and 2504 controls were conducted to investigate the association between the AS-SNPs and bladder cancer susceptibility. A series of assays were used to evaluate the functional effect of AS-SNPs on bladder cancer risk.
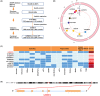
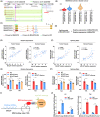
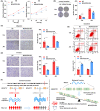
Results: We observed that SNP rs558814 A>G located in lncRNA BCLET (Bladder Cancer Low-Expressed Transcript, ENSG00000245498) can decrease the risk of bladder cancer (odds ratio [OR] = 0.84, 95% confidence interval [CI] = 0.76-0.92, p = 3.26 × 10-4 ). Additionally, the G allele of rs558814 had transcriptional regulatory effects and facilitated the expression of BCLET transcripts, including BCLET-long and BCLET-short. We also found decreased BCLET expression in bladder cancer tissues and cells, and BCLET transcript upregulation substantially inhibited tumor growth of both bladder cancer cells and xenograft models. Mechanistically, BCLET recognized and regulated AS of MSANTD2 to participate in bladder carcinogenesis, preferentially promoting the production of MSANTD2-004.
Conclusions: SNP rs558814 was associated with the expression of BCLET, which mainly increased the expression of MSANTD2-004 through AS of MSANTD2.
Keywords: alternative splicing; bladder cancer; genetic variations; lncRNA BCLET; molecular mechanism.
© 2023 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
No potential conflicts of interest are disclosed.
Figures


Similar articles
-
Alternative splicing related genetic variants contribute to bladder cancer risk.Mol Carcinog. 2020 Aug;59(8):923-929. doi: 10.1002/mc.23207. Epub 2020 Apr 27. Mol Carcinog. 2020. PMID: 32339354
-
lncRNA TINCR SNPs and Expression Levels Are Associated with Bladder Cancer Susceptibility.Genet Test Mol Biomarkers. 2021 Jan;25(1):31-41. doi: 10.1089/gtmb.2020.0178. Epub 2020 Dec 28. Genet Test Mol Biomarkers. 2021. PMID: 33372851 Clinical Trial.
-
Association between lncRNA H19 (rs217727, rs2735971 and rs3024270) polymorphisms and the risk of bladder cancer in Chinese population.Minerva Urol Nefrol. 2019 Apr;71(2):161-167. doi: 10.23736/S0393-2249.18.03004-7. Epub 2018 Mar 28. Minerva Urol Nefrol. 2019. PMID: 29595036
-
Genetic variants in splicing factor genes and susceptibility to bladder cancer.Gene. 2022 Jan 30;809:146022. doi: 10.1016/j.gene.2021.146022. Epub 2021 Oct 18. Gene. 2022. PMID: 34673209
-
Association between five polymorphisms in vascular endothelial growth factor gene and urinary bladder cancer risk: A systematic review and meta-analysis involving 6671 subjects.Gene. 2019 May 25;698:186-197. doi: 10.1016/j.gene.2019.02.070. Epub 2019 Mar 5. Gene. 2019. PMID: 30849545
Cited by
-
The implications for urological malignancies of non-coding RNAs in the the tumor microenvironment.Comput Struct Biotechnol J. 2023 Dec 20;23:491-505. doi: 10.1016/j.csbj.2023.12.016. eCollection 2024 Dec. Comput Struct Biotechnol J. 2023. PMID: 38249783 Free PMC article. Review.
References
-
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2022. CA Cancer J Clin. 2022;72(1):7‐33. - PubMed
-
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115‐132. - PubMed
-
- Burger M, Catto JW, Dalbagni G, et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol. 2013;63(2):234‐241. - PubMed
-
- Islami F, Goding Sauer A, Miller KD, et al. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J Clin. 2018;68(1):31‐54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

