The NO Answer for Autism Spectrum Disorder
- PMID: 37212048
- PMCID: PMC10401098
- DOI: 10.1002/advs.202205783
The NO Answer for Autism Spectrum Disorder
Abstract
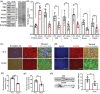
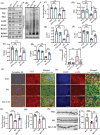
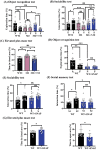
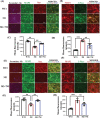
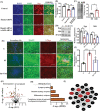
Autism spectrum disorders (ASDs) include a wide range of neurodevelopmental disorders. Several reports showed that mutations in different high-risk ASD genes lead to ASD. However, the underlying molecular mechanisms have not been deciphered. Recently, they reported a dramatic increase in nitric oxide (NO) levels in ASD mouse models. Here, they conducted a multidisciplinary study to investigate the role of NO in ASD. High levels of nitrosative stress biomarkers are found in both the Shank3 and Cntnap2 ASD mouse models. Pharmacological intervention with a neuronal NO synthase (nNOS) inhibitor in both models led to a reversal of the molecular, synaptic, and behavioral ASD-associated phenotypes. Importantly, treating iPSC-derived cortical neurons from patients with SHANK3 mutation with the nNOS inhibitor showed similar therapeutic effects. Clinically, they found a significant increase in nitrosative stress biomarkers in the plasma of low-functioning ASD patients. Bioinformatics of the SNO-proteome revealed that the complement system is enriched in ASD. This novel work reveals, for the first time, that NO plays a significant role in ASD. Their important findings will open novel directions to examine NO in diverse mutations on the spectrum as well as in other neurodevelopmental disorders. Finally, it suggests a novel strategy for effectively treating ASD.
Keywords: S-nitrosylation; Shank3; autism spectrum disorder; behavior; contactin-associated protein-like2; nitric oxide.
© 2023 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Christensen D. L., Braun K. V. N., Baio J., Bilder D., Charles J., Constantino J. N., Daniels J., Durkin M. S., Fitzgerald R. T., Kurzius‐Spencer M., Lee L.‐C., Pettygrove S., Robinson C., Schulz E., Wells C., Wingate M. S., Zahorodny W., Yeargin‐Allsopp M., MMWR Surveill. Summ. 2018, 65, 1. - PMC - PubMed
-
- Sharma S. R., Gonda X., Tarazi F. I., Pharmacol. Ther. 2018, 190, 91. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
