Information-seeking preferences in diverse patients receiving a genetic testing result in the Clinical Sequencing Evidence-Generating Research (CSER) study
- PMID: 37212252
- PMCID: PMC10524447
- DOI: 10.1016/j.gim.2023.100899
Information-seeking preferences in diverse patients receiving a genetic testing result in the Clinical Sequencing Evidence-Generating Research (CSER) study
Abstract
Purpose: Accurate and understandable information after genetic testing is critical for patients, family members, and professionals alike.
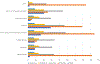
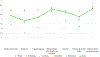
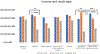
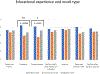
Methods: As part of a cross-site study from the Clinical Sequencing Evidence-Generating Research consortium, we investigated the information-seeking practices among patients and family members at 5 to 7 months after genetic testing results disclosure, assessing the perceived utility of a variety of information sources, such as family and friends, health care providers, support groups, and the internet.
Results: We found that individuals placed a high value on information obtained from genetics professionals and health care workers, independent of genetic testing result case classifications as positive, inconclusive, or negative. The internet was also highly utilized and ranked. Study participants rated some information sources as more useful for positive results compared with inconclusive or negative outcomes, emphasizing that it may be difficult to identify helpful information for individuals receiving an uncertain or negative result. There were few data from non-English speakers, highlighting the need to develop strategies to reach this population.
Conclusion: Our study emphasizes the need for clinicians to provide accurate and comprehensible information to individuals from diverse populations after genetic testing.
Keywords: Diverse populations; Exome sequencing; Genetic testing; Genome sequencing; Information-seeking preferences.
Copyright © 2023 American College of Medical Genetics and Genomics. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of Interest The authors declare no conflicts of interest.
Figures

References
-
- Stark Z, Tan TY, Chong B, Brett GR, Yap P, Walsh M, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med. 2016;18:1090–1096. - PubMed
-
- Zimmermann BM, Fanderl J, Koné I, Rabaglio M, Bürki N, Shaw D, Elger B. Examining information-seeking behavior in genetic testing for cancer predisposition: A qualitative interview study. Patient Educ Couns. 2021. Feb;104(2):257–264. - PubMed
-
- Jacobs W, Amuta AO, Chan Jeon K. Health information seeking in the digital age: An analysis of health information seeking behavior among US adults. Cogent Social Sciences 2017: 3: 1302785.
-
- Chavarria EA, Christy SM, Feng H, Miao H, Abdulla R, Gutierrez L, Lopez D, Sanchez J, Gwede CK, Meade CD. Online Health Information Seeking and eHealth Literacy Among Spanish Language-Dominant Latino Adults Receiving Care in a Community Clinic: Secondary Analysis of Pilot Randomized Controlled Trial Data. JMIR Form Res. 2022. Oct 13;6(10):e37687. doi: 10.2196/37687. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

