Hypothalamic and brainstem glucose-dependent insulinotropic polypeptide receptor neurons employ distinct mechanisms to affect feeding
- PMID: 37212283
- PMCID: PMC10322681
- DOI: 10.1172/jci.insight.164921
Hypothalamic and brainstem glucose-dependent insulinotropic polypeptide receptor neurons employ distinct mechanisms to affect feeding
Abstract
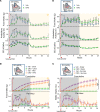
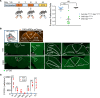
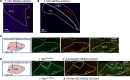
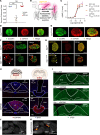
Central glucose-dependent insulinotropic polypeptide (GIP) receptor (GIPR) signaling is critical in GIP-based therapeutics' ability to lower body weight, but pathways leveraged by GIPR pharmacology in the brain remain incompletely understood. We explored the role of Gipr neurons in the hypothalamus and dorsal vagal complex (DVC) - brain regions critical to the control of energy balance. Hypothalamic Gipr expression was not necessary for the synergistic effect of GIPR/GLP-1R coagonism on body weight. While chemogenetic stimulation of both hypothalamic and DVC Gipr neurons suppressed food intake, activation of DVC Gipr neurons reduced ambulatory activity and induced conditioned taste avoidance, while there was no effect of a short-acting GIPR agonist (GIPRA). Within the DVC, Gipr neurons of the nucleus tractus solitarius (NTS), but not the area postrema (AP), projected to distal brain regions and were transcriptomically distinct. Peripherally dosed fluorescent GIPRAs revealed that access was restricted to circumventricular organs in the CNS. These data demonstrate that Gipr neurons in the hypothalamus, AP, and NTS differ in their connectivity, transcriptomic profile, peripheral accessibility, and appetite-controlling mechanisms. These results highlight the heterogeneity of the central GIPR signaling axis and suggest that studies into the effects of GIP pharmacology on feeding behavior should consider the interplay of multiple regulatory pathways.
Keywords: Diabetes; Metabolism; Neuroendocrine regulation; Neuroscience; Obesity.
Conflict of interest statement
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
- MC_UU_12012/3/MRC_/Medical Research Council/United Kingdom
- MR/N00275X/1/MRC_/Medical Research Council/United Kingdom
- 220271/Z/20/Z/WT_/Wellcome Trust/United Kingdom
- R01 DK124276/DK/NIDDK NIH HHS/United States
- MC_UU_00014/3/MRC_/Medical Research Council/United Kingdom
- MR/W000881/2/MRC_/Medical Research Council/United Kingdom
- R01 DK125353/DK/NIDDK NIH HHS/United States
- 223279/Z/21/Z/WT_/Wellcome Trust/United Kingdom
- K01 DK132461/DK/NIDDK NIH HHS/United States
- R37 DK046492/DK/NIDDK NIH HHS/United States
- R01 DK123075/DK/NIDDK NIH HHS/United States
- WT_/Wellcome Trust/United Kingdom
- MR/S025618/1/MRC_/Medical Research Council/United Kingdom
- MRC_MC_UU_12012/3/MRC_/Medical Research Council/United Kingdom
- MR/W000881/1/MRC_/Medical Research Council/United Kingdom
- R01 DK046492/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

