Honokiol Inhibits SARS-CoV-2 Replication in Cell Culture at a Post-Entry Step
- PMID: 37212560
- PMCID: PMC10269499
- DOI: 10.1128/spectrum.03273-22
Honokiol Inhibits SARS-CoV-2 Replication in Cell Culture at a Post-Entry Step
Abstract
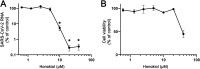
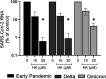
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in 2019, and the resulting pandemic has already caused the death of over 6 million people. There are currently few antivirals approved for treatment of the 2019 coronavirus disease (COVID-19), and more options would be beneficial, not only now but also to increase our preparedness for future coronavirus outbreaks. Honokiol is a small molecule from magnolia trees for which several biological effects have been reported, including anticancer and anti-inflammatory activities. Honokiol has also been shown to inhibit several viruses in cell culture. In this study, we determined that honokiol protected Vero E6 cells from SARS-CoV-2-mediated cytopathic effect, with a 50% effective concentration of 7.8 μM. In viral load reduction assays, honokiol decreased viral RNA copies as well as viral infectious progeny titers. The compound also inhibited SARS-CoV-2 replication in the more relevant human A549 cells expressing angiotensin converting enzyme 2 and transmembrane protease serine 2. Time-of-addition and other assays showed that honokiol inhibited virus replication at a post-entry step of the replication cycle. Honokiol was also effective against more recent variants of SARS-CoV-2, including Omicron, and it inhibited other human coronaviruses as well. Our study suggests that honokiol is an interesting molecule to be evaluated further in animal studies and, when successful, maybe even in clinical trials to investigate its effect on virus replication and pathogenic (inflammatory) host responses. IMPORTANCE Honokiol is a compound that shows both anti-inflammatory and antiviral effects, and therefore its effect on SARS-CoV-2 infection was assessed. This small molecule inhibited SARS-CoV-2 replication in various cell-based infection systems, with up to an ~1,000-fold reduction in virus titer. In contrast to earlier reports, our study clearly showed that honokiol acts on a postentry step of the replication cycle. Honokiol also inhibited different recent SARS-CoV-2 variants and other human coronaviruses (Middle East respiratory syndrome CoV and SARS-CoV), demonstrating its broad spectrum of antiviral activity. The anticoronavirus effect, combined with its anti-inflammatory properties, make honokiol an interesting compound to be further explored in animal coronavirus infection models.
Keywords: MERS-CoV; SARS-CoV; SARS-CoV-2; antiviral agents; coronaviruses; honokiol; inhibitor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

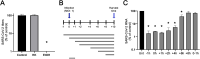


References
-
- Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. 2020. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395:497–506. doi:10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
-
- Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, Datir R, Collier DA, Albecka A, Singh S, Pandey R, Brown J, Zhou J, Goonawardane N, Mishra S, Whittaker C, Mellan T, Marwal R, Datta M, Sengupta S, Ponnusamy K, Radhakrishnan VS, Abdullahi A, Charles O, Chattopadhyay P, Devi P, Caputo D, Peacock T, Wattal C, Goel N, Satwik A, Vaishya R, Agarwal M, Indian SARS-COV-2 Genomics Consortium, Genotype to Phenotype Japan (G2P-Japan) Consortium, CITIID-NIHR BioResource COVID-19 Collaboration, Mavousian A, Lee JH, Bassi J, Silacci-Fegni C, Saliba C, Pinto D, Irie T, Yoshida I, Hamilton WL, Sato K, Bhatt S, Flaxman S, James LC, Corti D, et al.. 2021. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599:114–119. doi:10.1038/s41586-021-03944-y. - DOI - PMC - PubMed
-
- Cheng VCC, Ip JD, Chu AWH, Tam AR, Chan WM, Abdullah SMU, Chan BPC, Wong SC, Kwan MYW, Chua GT, Ip P, Chan JMC, Lam BHS, To WK, Chuang VWM, Yuen KY, Hung IFN, To KKW. 2022. Rapid spread of SARS-CoV-2 Omicron subvariant BA.2 in a single-source community outbreak. Clin Infect Dis 75:e44–e49. doi:10.1093/cid/ciac203. - DOI - PMC - PubMed
-
- Ong SWX, Chiew CJ, Ang LW, Mak TM, Cui L, Toh M, Lim YD, Lee PH, Lee TH, Chia PY, Maurer-Stroh S, Lin RTP, Leo YS, Lee VJ, Lye DC, Young BE. 2021. Clinical and virological features of SARS-CoV-2 variants of concern: a retrospective cohort study comparing B.1.1.7 (Alpha), B.1.315 (Beta), and B.1.617.2 (Delta). Clin Infect Dis 75:e1128–e1136. doi:10.1093/cid/ciab721. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

