Rhizosphere microbial community composition shifts diurnally and in response to natural variation in host clock phenotype
- PMID: 37212579
- PMCID: PMC10308896
- DOI: 10.1128/msystems.01487-21
Rhizosphere microbial community composition shifts diurnally and in response to natural variation in host clock phenotype
Abstract
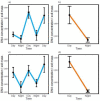
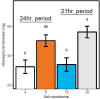
Plant-associated microbial assemblages are known to shift at time scales aligned with plant phenology, as influenced by the changes in plant-derived nutrient concentrations and abiotic conditions observed over a growing season. But these same factors can change dramatically in a sub-24-hour period, and it is poorly understood how such diel cycling may influence plant-associated microbiomes. Plants respond to the change from day to night via mechanisms collectively referred to as the internal "clock," and clock phenotypes are associated with shifts in rhizosphere exudates and other changes that we hypothesize could affect rhizosphere microbes. The mustard Boechera stricta has wild populations that contain multiple clock phenotypes of either a 21- or a 24-hour cycle. We grew plants of both phenotypes (two genotypes per phenotype) in incubators that simulated natural diel cycling or that maintained constant light and temperature. Under both cycling and constant conditions, the extracted DNA concentration and the composition of rhizosphere microbial assemblages differed between time points, with daytime DNA concentrations often triple what were observed at night and microbial community composition differing by, for instance, up to 17%. While we found that plants of different genotypes were associated with variation in rhizosphere assemblages, we did not see an effect on soil conditioned by a particular host plant circadian phenotype on subsequent generations of plants. Our results suggest that rhizosphere microbiomes are dynamic at sub-24-hour periods, and those dynamics are shaped by diel cycling in host plant phenotype. IMPORTANCE We find that the rhizosphere microbiome shifts in composition and extractable DNA concentration in sub-24-hour periods as influenced by the plant host's internal clock. These results suggest that host plant clock phenotypes could be an important determinant of variation in rhizosphere microbiomes.
Keywords: diel cylcing; dynamics; microbiome; rhizosphere.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Jones DL, Nguyen C, Finlay RD. 2009. Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321:5–33. doi:10.1007/s11104-009-9925-0 - DOI
-
- Sperry JS, Adler FR, Campbell GS, Comstock JP. 1998. Limitation of plant water use by rhizosphere and xylem conductance: results from a model. Plant Cell Environ 21:347–359. doi:10.1046/j.1365-3040.1998.00287.x - DOI
-
- Huang X-F, Chaparro JM, Reardon KF, Zhang R, Shen Q, Vivanco JM. 2014. Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92:267–275. doi:10.1139/cjb-2013-0225 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
