Distinct Epithelial-Innate Immune Cell Transcriptional Circuits Underlie Airway Hyperresponsiveness in Asthma
- PMID: 37212596
- PMCID: PMC10273121
- DOI: 10.1164/rccm.202209-1707OC
Distinct Epithelial-Innate Immune Cell Transcriptional Circuits Underlie Airway Hyperresponsiveness in Asthma
Abstract
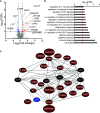
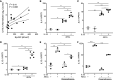
Rationale: Indirect airway hyperresponsiveness (AHR) is a highly specific feature of asthma, but the underlying mechanisms responsible for driving indirect AHR remain incompletely understood. Objectives: To identify differences in gene expression in epithelial brushings obtained from individuals with asthma who were characterized for indirect AHR in the form of exercise-induced bronchoconstriction (EIB). Methods: RNA-sequencing analysis was performed on epithelial brushings obtained from individuals with asthma with EIB (n = 11) and without EIB (n = 9). Differentially expressed genes (DEGs) between the groups were correlated with measures of airway physiology, sputum inflammatory markers, and airway wall immunopathology. On the basis of these relationships, we examined the effects of primary airway epithelial cells (AECs) and specific epithelial cell-derived cytokines on both mast cells (MCs) and eosinophils (EOS). Measurements and Main Results: We identified 120 DEGs in individuals with and without EIB. Network analyses suggested critical roles for IL-33-, IL-18-, and IFN-γ-related signaling among these DEGs. IL1RL1 expression was positively correlated with the density of MCs in the epithelial compartment, and IL1RL1, IL18R1, and IFNG were positively correlated with the density of intraepithelial EOS. Subsequent ex vivo modeling demonstrated that AECs promote sustained type 2 (T2) inflammation in MCs and enhance IL-33-induced T2 gene expression. Furthermore, EOS increase the expression of IFNG and IL13 in response to both IL-18 and IL-33 as well as exposure to AECs. Conclusions: Circuits involving epithelial interactions with MCs and EOS are closely associated with indirect AHR. Ex vivo modeling indicates that epithelial-dependent regulation of these innate cells may be critical in indirect AHR and modulating T2 and non-T2 inflammation in asthma.
Keywords: airway epithelium; airway hyperresponsiveness; asthma; eosinophil; mast cell.
Figures


Comment in
-
The ABCs and DEGs (Differentially Expressed Genes) of Airway Hyperresponsiveness.Am J Respir Crit Care Med. 2023 Jun 15;207(12):1545-1546. doi: 10.1164/rccm.202303-0614ED. Am J Respir Crit Care Med. 2023. PMID: 37058325 Free PMC article. No abstract available.
References
-
- Hallstrand TS, Leuppi JD, Joos G, Hall GL, Carlsen KH, Kaminsky DA, et al. American Thoracic Society (ATS)/European Respiratory Society (ERS) Bronchoprovocation Testing Task Force ERS technical standard on bronchial challenge testing: pathophysiology and methodology of indirect airway challenge testing. Eur Respir J . 2018;52:1801033. - PubMed
-
- Coates AL, Wanger J, Cockcroft DW, Culver BH, Carlsen K-H, Diamant Z, et al. Bronchoprovocation Testing Task Force ERS technical standard on bronchial challenge testing: general considerations and performance of methacholine challenge tests. Eur Respir J . 2017;49:1601526. - PubMed
-
- Cabral AL, Conceição GM, Fonseca-Guedes CH, Martins MA. Exercise-induced bronchospasm in children: effects of asthma severity. Am J Respir Crit Care Med . 1999;159:1819–1823. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

