The structural analysis of the periplasmic domain of Sinorhizobium meliloti chemoreceptor McpZ reveals a novel fold and suggests a complex mechanism of transmembrane signaling
- PMID: 37213073
- PMCID: PMC10524373
- DOI: 10.1002/prot.26510
The structural analysis of the periplasmic domain of Sinorhizobium meliloti chemoreceptor McpZ reveals a novel fold and suggests a complex mechanism of transmembrane signaling
Abstract
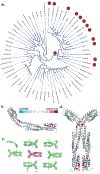
Chemotaxis is a fundamental process whereby bacteria seek out nutrient sources and avoid harmful chemicals. For the symbiotic soil bacterium Sinorhizobium meliloti, the chemotaxis system also plays an essential role in the interaction with its legume host. The chemotactic signaling cascade is initiated through interactions of an attractant or repellent compound with chemoreceptors or methyl-accepting chemotaxis proteins (MCPs). S. meliloti possesses eight chemoreceptors to mediate chemotaxis. Six of these receptors are transmembrane proteins with periplasmic ligand-binding domains (LBDs). The specific functions of McpW and McpZ are still unknown. Here, we report the crystal structure of the periplasmic domain of McpZ (McpZPD) at 2.7 Å resolution. McpZPD assumes a novel fold consisting of three concatenated four-helix bundle modules. Through phylogenetic analyses, we discovered that this helical tri-modular domain fold arose within the Rhizobiaceae family and is still evolving rapidly. The structure, offering a rare view of a ligand-free dimeric MCP-LBD, reveals a novel dimerization interface. Molecular dynamics calculations suggest ligand binding will induce conformational changes that result in large horizontal helix movements within the membrane-proximal domains of the McpZPD dimer that are accompanied by a 5 Å vertical shift of the terminal helix toward the inner cell membrane. These results suggest a mechanism of transmembrane signaling for this family of MCPs that entails both piston-type and scissoring movements. The predicted movements terminate in a conformation that closely mirrors those observed in related ligand-bound MCP-LBDs.
Keywords: chemotaxis; helical tri-modular sensor domain; ligand-binding domain; methyl-accepting chemotaxis protein; piston; scissoring; transmembrane signaling.
© 2023 The Authors. Proteins: Structure, Function, and Bioinformatics published by Wiley Periodicals LLC.
Figures

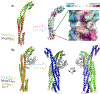
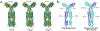
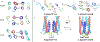
Similar articles
-
Cellular Stoichiometry of Methyl-Accepting Chemotaxis Proteins in Sinorhizobium meliloti.J Bacteriol. 2018 Feb 23;200(6):e00614-17. doi: 10.1128/JB.00614-17. Print 2018 Mar 15. J Bacteriol. 2018. PMID: 29263102 Free PMC article.
-
Functional analysis of nine putative chemoreceptor proteins in Sinorhizobium meliloti.J Bacteriol. 2007 Mar;189(5):1816-26. doi: 10.1128/JB.00883-06. Epub 2006 Dec 22. J Bacteriol. 2007. PMID: 17189365 Free PMC article.
-
The ligand-binding domain of a chemoreceptor from Comamonas testosteroni has a previously unknown homotrimeric structure.Mol Microbiol. 2019 Sep;112(3):906-917. doi: 10.1111/mmi.14326. Epub 2019 Jun 21. Mol Microbiol. 2019. PMID: 31177588 Free PMC article.
-
Sensory transduction to the flagellar motor of Sinorhizobium meliloti.J Mol Microbiol Biotechnol. 2002 May;4(3):183-6. J Mol Microbiol Biotechnol. 2002. PMID: 11931544 Review.
-
Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea.Cell Mol Life Sci. 2017 Sep;74(18):3293-3303. doi: 10.1007/s00018-017-2514-0. Epub 2017 Apr 13. Cell Mol Life Sci. 2017. PMID: 28409190 Free PMC article. Review.
Cited by
-
Bacterial sensor evolved by decreasing complexity.Proc Natl Acad Sci U S A. 2025 Feb 4;122(5):e2409881122. doi: 10.1073/pnas.2409881122. Epub 2025 Jan 29. Proc Natl Acad Sci U S A. 2025. PMID: 39879239 Free PMC article.
-
Structural and functional diversity of sensor domains in bacterial transmembrane receptors.Trends Microbiol. 2025 Jul;33(7):796-809. doi: 10.1016/j.tim.2025.02.019. Epub 2025 Mar 21. Trends Microbiol. 2025. PMID: 40121131 Review.
-
The motility and chemosensory systems of Rhizobium leguminosarum, their role in symbiosis, and link to PTSNtr regulation.Environ Microbiol. 2024 Feb;26(2):e16570. doi: 10.1111/1462-2920.16570. Epub 2024 Jan 12. Environ Microbiol. 2024. PMID: 38216524 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

