In Silico Identification and Analysis of Potentially Bioactive Antiviral Phytochemicals against SARS-CoV-2: A Molecular Docking and Dynamics Simulation Approach
- PMID: 37214084
- PMCID: PMC10195178
- DOI: 10.1155/2023/5469258
In Silico Identification and Analysis of Potentially Bioactive Antiviral Phytochemicals against SARS-CoV-2: A Molecular Docking and Dynamics Simulation Approach
Abstract
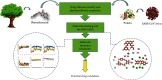
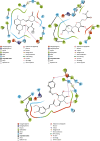
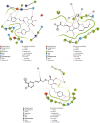
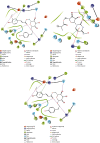
SARS-CoV-2, a deadly coronavirus sparked COVID-19 pandemic around the globe. With an increased mutation rate, this infectious agent is highly transmissible inducing an escalated rate of infections and death everywhere. Hence, the discovery of a viable antiviral therapy option is urgent. Computational approaches have offered a revolutionary framework to identify novel antimicrobial treatment regimens and allow a quicker, cost-effective, and productive conversion into the health center by evaluating preliminary and safety investigations. The primary purpose of this research was to find plausible plant-derived antiviral small molecules to halt the viral entrance into individuals by clogging the adherence of Spike protein with human ACE2 receptor and to suppress their genome replication by obstructing the activity of Nsp3 (Nonstructural protein 3) and 3CLpro (main protease). An in-house library of 1163 phytochemicals were selected from the NPASS and PubChem databases for downstream analysis. Preliminary analysis with SwissADME and pkCSM revealed 149 finest small molecules from the large dataset. Virtual screening using the molecular docking scoring and the MM-GBSA data analysis revealed that three candidate ligands CHEMBL503 (Lovastatin), CHEMBL490355 (Sulfuretin), and CHEMBL4216332 (Grayanoside A) successfully formed docked complex within the active site of human ACE2 receptor, Nsp3, and 3CLpro, respectively. Dual method molecular dynamics (MD) simulation and post-MD MM-GBSA further confirmed efficient binding and stable interaction between the ligands and target proteins. Furthermore, biological activity spectra and molecular target analysis revealed that all three preselected phytochemicals were biologically active and safe for human use. Throughout the adopted methodology, all three therapeutic candidates significantly outperformed the control drugs (Molnupiravir and Paxlovid). Finally, our research implies that these SARS-CoV-2 protein antagonists might be viable therapeutic options. At the same time, enough wet lab evaluations would be needed to ensure the therapeutic potency of the recommended drug candidates for SARS-CoV-2.
Copyright © 2023 Sajal Kumar Halder et al.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures









References
-
- Shawan M. M. A. K., Halder S. K., Hasan M. A. Luteolin and abyssinone II as potential inhibitors of SARS-CoV-2: an in silico molecular modeling approach in battling the COVID-19 outbreak. Bulletin of the National Research Centre . 2021;45(1):p. 27. doi: 10.1186/s42269-020-00479-6. - DOI - PMC - PubMed
-
- WHO Coronavirus (COVID-19) Dashboard | WHO Coronavirus (COVID-19) Dashboard with Vaccination Data. https://covid19.who.int/
-
- Srivastava A. K., Kumar A., Misra N. On the inhibition of COVID-19 protease by Indian herbal plants: an in silico investigation. 2020, https://arxiv.org/abs/2004.03411.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

