Functional and comparative analysis of THI1 gene in grasses with a focus on sugarcane
- PMID: 37214086
- PMCID: PMC10194071
- DOI: 10.7717/peerj.14973
Functional and comparative analysis of THI1 gene in grasses with a focus on sugarcane
Abstract
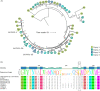
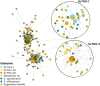
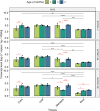
De novo synthesis of thiamine (vitamin B1) in plants depends on the action of thiamine thiazole synthase, which synthesizes the thiazole ring, and is encoded by the THI1 gene. Here, we investigated the evolution and diversity of THI1 in Poaceae, where C4 and C3 photosynthetic plants co-evolved. An ancestral duplication of THI1 is observed in Panicoideae that remains in many modern monocots, including sugarcane. In addition to the two sugarcane copies (ScTHI1-1 and ScTHI1-2), we identified ScTHI1-2 alleles showing differences in their sequence, indicating divergence between ScTHI1-2a and ScTHI1-2b. Such variations are observed only in the Saccharum complex, corroborating the phylogeny. At least five THI1 genomic environments were found in Poaceae, two in sugarcane, M. sinensis, and S. bicolor. The THI1 promoter in Poaceae is highly conserved at 300 bp upstream of the start codon ATG and has cis-regulatory elements that putatively bind to transcription factors associated with development, growth, development and biological rhythms. An experiment set to compare gene expression levels in different tissues across the sugarcane R570 life cycle showed that ScTHI1-1 was expressed mainly in leaves regardless of age. Furthermore, ScTHI1 displayed relatively high expression levels in meristem and culm, which varied with the plant age. Finally, yeast complementation studies with THI4-defective strain demonstrate that only ScTHI1-1 and ScTHI1-2b isoforms can partially restore thiamine auxotrophy, albeit at a low frequency. Taken together, the present work supports the existence of multiple origins of THI1 harboring genomic regions in Poaceae with predicted functional redundancy. In addition, it questions the contribution of the levels of the thiazole ring in C4 photosynthetic plant tissues or potentially the relevance of the THI1 protein activity.
Keywords: Evolutionary diversity; Gene expression; Genetic complementation; Genomic characterization; Plant development; Promoter analysis; Thiazole biosynthesis.
© 2023 Moura Dias et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

