This is a preprint.
A deep catalog of protein-coding variation in 985,830 individuals
- PMID: 37214792
- PMCID: PMC10197621
- DOI: 10.1101/2023.05.09.539329
A deep catalog of protein-coding variation in 985,830 individuals
Abstract
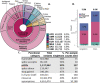
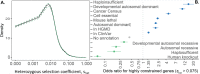
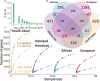
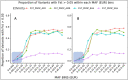
Coding variants that have significant impact on function can provide insights into the biology of a gene but are typically rare in the population. Identifying and ascertaining the frequency of such rare variants requires very large sample sizes. Here, we present the largest catalog of human protein-coding variation to date, derived from exome sequencing of 985,830 individuals of diverse ancestry to serve as a rich resource for studying rare coding variants. Individuals of African, Admixed American, East Asian, Middle Eastern, and South Asian ancestry account for 20% of this Exome dataset. Our catalog of variants includes approximately 10.5 million missense (54% novel) and 1.1 million predicted loss-of-function (pLOF) variants (65% novel, 53% observed only once). We identified individuals with rare homozygous pLOF variants in 4,874 genes, and for 1,838 of these this work is the first to document at least one pLOF homozygote. Additional insights from the RGC-ME dataset include 1) improved estimates of selection against heterozygous loss-of-function and identification of 3,459 genes intolerant to loss-of-function, 83 of which were previously assessed as tolerant to loss-of-function and 1,241 that lack disease annotations; 2) identification of regions depleted of missense variation in 457 genes that are tolerant to loss-of-function; 3) functional interpretation for 10,708 variants of unknown or conflicting significance reported in ClinVar as cryptic splice sites using splicing score thresholds based on empirical variant deleteriousness scores derived from RGC-ME; and 4) an observation that approximately 3% of sequenced individuals carry a clinically actionable genetic variant in the ACMG SF 3.1 list of genes. We make this important resource of coding variation available to the public through a variant allele frequency browser. We anticipate that this report and the RGC-ME dataset will serve as a valuable reference for understanding rare coding variation and help advance precision medicine efforts.
Figures


References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
