This is a preprint.
IFNα induces CCR5 in CD4+ T-cells, causing its anti- HIV inefficiency and its subsequent pathogenic elevation, partially controlled by anti-HIV therapy
- PMID: 37214795
- PMCID: PMC10197818
- DOI: 10.21203/rs.3.rs-2813616/v1
IFNα induces CCR5 in CD4+ T-cells, causing its anti- HIV inefficiency and its subsequent pathogenic elevation, partially controlled by anti-HIV therapy
Update in
-
IFNα induces CCR5 in CD4+ T cells of HIV patients causing pathogenic elevation.Commun Med (Lond). 2024 Mar 19;4(1):52. doi: 10.1038/s43856-024-00453-7. Commun Med (Lond). 2024. PMID: 38504093 Free PMC article.
Abstract
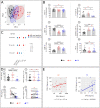
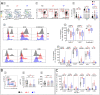
Like EC, we find that ART-treated patients control serum IFNα concentration and show few immune cell alterations enabling a healthy but fragile medical status. However, treatment interruption leads to elevated IFNα reflecting virus production indicating that like EC, ART does not achieve a virological cure. The immune system becomes overwhelmed by multiple immune cell abnormalities as found in untreated patients. These are chiefly mediated by elevated IFNα inducing signaling checkpoints abnormalities, including PD1, in cytotoxic immune cells. Importantly, during acute infection, elevated IFNα correlated with HIV load and we found that IFNα enhances CCR5, the HIV coreceptor in CD4+ T-cells, impairing its anti-viral response and accounting for the pathogenic vicious cycle: HIV → IFNα ↗ → infected CD4+ T-cells ↗ →HIV ↗. This study opens immunotherapeutic perspectives showing the need to control IFNα in order to convert ART patients into EC.
Conflict of interest statement
Additional Declarations: There is NO Competing Interest.
Figures







References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

