This is a preprint.
Deletion of miR-146a enhances therapeutic protein restoration in model of dystrophin exon skipping
- PMID: 37214870
- PMCID: PMC10197665
- DOI: 10.1101/2023.05.09.540042
Deletion of miR-146a enhances therapeutic protein restoration in model of dystrophin exon skipping
Update in
-
Deletion of miR-146a enhances therapeutic protein restoration in model of dystrophin exon skipping.Mol Ther Nucleic Acids. 2024 May 24;35(3):102228. doi: 10.1016/j.omtn.2024.102228. eCollection 2024 Sep 10. Mol Ther Nucleic Acids. 2024. PMID: 38975000 Free PMC article.
Abstract
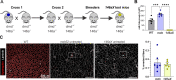
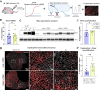
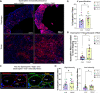
Duchenne muscular dystrophy (DMD) is a progressive muscle disease caused by the absence of dystrophin protein. One current DMD therapeutic strategy, exon skipping, produces a truncated dystrophin isoform using phosphorodiamidate morpholino oligomers (PMOs). However, the potential of exon skipping therapeutics has not been fully realized as increases in dystrophin protein have been minimal in clinical trials. Here, we investigate how miR-146a-5p, which is highly elevated in dystrophic muscle, impacts dystrophin protein levels. We find inflammation strongly induces miR-146a in dystrophic, but not wild-type myotubes. Bioinformatics analysis reveals that the dystrophin 3'UTR harbors a miR-146a binding site, and subsequent luciferase assays demonstrate miR-146a binding inhibits dystrophin translation. In dystrophin-null mdx52 mice, co-injection of miR-146a reduces dystrophin restoration by an exon 51 skipping PMO. To directly investigate how miR-146a impacts therapeutic dystrophin rescue, we generated mdx52 with body-wide miR-146a deletion (146aX). Administration of an exon skipping PMO via intramuscular or intravenous injection markedly increases dystrophin protein levels in 146aX versus mdx52 muscles; skipped dystrophin transcript levels are unchanged, suggesting a post-transcriptional mechanism-of-action. Together, these data show that miR-146a expression opposes therapeutic dystrophin restoration, suggesting miR-146a inhibition warrants further research as a potential DMD exon skipping co-therapy.
Keywords: Becker Muscular Dystrophy (BMD); Duchenne Muscular Dystrophy (DMD); RNA therapeutics; antisense oligonucleotides; dystrophin; exon skipping; miR-146a-5p; microRNA.
Conflict of interest statement
Conflict of Interest AAF has an issued patent on intellectual property relating to the manuscript (United States Patent #10266824).
Figures




References
-
- Hoffman EP, Brown RH Jr., and Kunkel LM (1987). Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell 51: 919–928. - PubMed
-
- Hoffman EP, Kunkel LM, Angelini C, Clarke A, Johnson M, and Harris JB (1989). Improved diagnosis of Becker muscular dystrophy by dystrophin testing. Neurology 39: 1011–1017. - PubMed
-
- Kesari A, Pirra LN, Bremadesam L, McIntyre O, Gordon E, Dubrovsky AL, et al. (2008). Integrated DNA, cDNA, and protein studies in Becker muscular dystrophy show high exception to the reading frame rule. Hum Mutat 29: 728–737. - PubMed
-
- van den Bergen JC, Wokke BH, Janson AA, van Duinen SG, Hulsker MA, Ginjaar HB, et al. (2014). Dystrophin levels and clinical severity in Becker muscular dystrophy patients. J Neurol Neurosurg Psychiatry 85: 747–753. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
