This is a preprint.
Differences in gut metagenomes between dairy workers and community controls: a cross-sectional study
- PMID: 37215025
- PMCID: PMC10197731
- DOI: 10.1101/2023.05.10.540270
Differences in gut metagenomes between dairy workers and community controls: a cross-sectional study
Update in
-
A cross-sectional comparison of gut metagenomes between dairy workers and community controls.BMC Genomics. 2024 Jul 20;25(1):708. doi: 10.1186/s12864-024-10562-1. BMC Genomics. 2024. PMID: 39033279 Free PMC article.
Abstract
Background: As a nexus of routine antibiotic use and zoonotic pathogen presence, the livestock farming environment is a potential hotspot for the emergence of zoonotic diseases and antibiotic resistant bacteria. Livestock can further facilitate disease transmission by serving as intermediary hosts for pathogens as they undergo evolution prior to a spillover event. In light of this, we are interested in characterizing the microbiome and resistome of dairy workers, whose exposure to the livestock farming environment places them at risk for facilitating community transmission of antibiotic resistant genes and emerging zoonotic diseases.
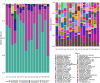
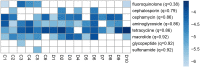
Results: Using shotgun sequencing, we investigated differences in the taxonomy, diversity and gene presence of the human gut microbiome of 10 dairy farm workers and 6 community controls, supplementing these samples with additional publicly available gut metagenomes. We observed greater abundance of tetracycline resistance genes and prevalence of cephamycin resistance genes in dairy workers' metagenomes, and lower average gene diversity. We also found evidence of commensal organism association with plasmid-mediated tetracycline resistance genes in both dairy workers and community controls (including Faecalibacterium prausnitzii, Ligilactobacillus animalis, and Simiaoa sunii). However, we did not find significant differences in the prevalence of resistance genes or virulence factors overall, nor differences in the taxonomic composition of dairy worker and community control metagenomes.
Conclusions: This study presents the first metagenomics analysis of United States dairy workers, providing insights into potential risks of exposure to antibiotics and pathogens in animal farming environments. Previous metagenomic studies of livestock workers in China and Europe have reported increased abundance and carriage of antibiotic resistance genes in livestock workers. While our investigation found no strong evidence for differences in the abundance or carriage of antibiotic resistance genes and virulence factors between dairy worker and community control gut metagenomes, we did observe patterns in the abundance of tetracycline resistance genes and the prevalence of cephamycin resistance genes that is consistent with previous work.
Figures

Similar articles
-
A cross-sectional comparison of gut metagenomes between dairy workers and community controls.BMC Genomics. 2024 Jul 20;25(1):708. doi: 10.1186/s12864-024-10562-1. BMC Genomics. 2024. PMID: 39033279 Free PMC article.
-
Airborne bacterial community and antibiotic resistome in the swine farming environment: Metagenomic insights into livestock relevance, pathogen hosts and public risks.Environ Int. 2023 Feb;172:107751. doi: 10.1016/j.envint.2023.107751. Epub 2023 Jan 13. Environ Int. 2023. PMID: 36680804
-
Description and determinants of the faecal resistome and microbiome of farmers and slaughterhouse workers: A metagenome-wide cross-sectional study.Environ Int. 2020 Oct;143:105939. doi: 10.1016/j.envint.2020.105939. Epub 2020 Jul 14. Environ Int. 2020. PMID: 32679392
-
Zoonotic bacteria in the vicinity of animal farms as a factor disturbing the human microbiome: a review.Int J Occup Med Environ Health. 2024 May 20;37(2):138-152. doi: 10.13075/ijomeh.1896.02003. Epub 2024 Apr 4. Int J Occup Med Environ Health. 2024. PMID: 38577723 Free PMC article. Review.
-
Invited review: Fate of antibiotic residues, antibiotic-resistant bacteria, and antibiotic resistance genes in US dairy manure management systems.J Dairy Sci. 2020 Feb;103(2):1051-1071. doi: 10.3168/jds.2019-16778. Epub 2019 Dec 16. J Dairy Sci. 2020. PMID: 31837779 Review.
References
-
- Abt M. C. and Pamer E. G.. Commensal bacteria mediated defenses against pathogens. Current Opinion in Immunology, 29:16–22, 2014. ISSN 0952–7915. doi: 10.1016/j.coi.2014.03.003. URL https://www.sciencedirect.com/science/article/pii/S095279151400048X. - DOI - PMC - PubMed
-
- Alcock B. P., Raphenya A. R., Lau T. T. Y., Tsang K. K., Bouchard M., Edalatmand A., Huynh W., Nguyen A.-L. V., Cheng A. A., Liu S., Min S. Y., Miroshnichenko A., Tran H.-K., Werfalli R. E., Nasir J. A., Oloni M., Speicher D. J., Florescu A., Singh B., Faltyn M., Hernandez-Koutoucheva A., Sharma A. N., Bordeleau E., Pawlowski A. C., Zubyk H. L., Dooley D., Griffiths E., Maguire F., Winsor G. L., Beiko R. G., Brinkman F. S. L., Hsiao W. W. L., Domselaar G. V., and McArthur A. G.. Card 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic acids research, 48(D1):D517–D525, 01 2020. doi: 10.1093/nar/gkz935. URL https://pubmed.ncbi.nlm.nih.gov/31665441. - DOI - PMC - PubMed
-
- Arrieta M.-C. and Finlay B.. The commensal microbiota drives immune homeostasis. Frontiers in Immunology, 3, 2012. ISSN 1664–3224. doi: 10.3389/fimmu.2012.00033. URL https://www.frontiersin.org/article/10.3389/fimmu.2012.00033. - DOI - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
