This is a preprint.
TGF-β Ligand Cross-Subfamily Interactions in the Response of Caenorhabditis elegans to Bacterial Pathogens
- PMID: 37215035
- PMCID: PMC10197529
- DOI: 10.1101/2023.05.05.539606
TGF-β Ligand Cross-Subfamily Interactions in the Response of Caenorhabditis elegans to Bacterial Pathogens
Update in
-
TGF-β ligand cross-subfamily interactions in the response of Caenorhabditis elegans to a bacterial pathogen.PLoS Genet. 2024 Jun 14;20(6):e1011324. doi: 10.1371/journal.pgen.1011324. eCollection 2024 Jun. PLoS Genet. 2024. PMID: 38875298 Free PMC article.
Abstract
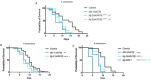
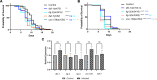
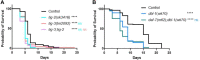
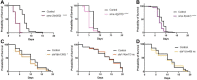
The Transforming Growth Factor beta (TGF-β) family consists of numerous secreted peptide growth factors that play significant roles in cell function, tissue patterning, and organismal homeostasis, including wound repair and immunity. Typically studied as homodimers, these ligands have the potential to diversify their functions through ligand interactions that are synergistic, cooperative, additive, and/or antagonistic. In the nematode Caenorhabditis elegans, there are only five TGF-β ligands, providing an opportunity to dissect ligand interactions in fewer combinations than in vertebrates. As in vertebrates, these ligands can be divided into bone morphogenetic protein (BMP) and TGF-β/Activin subfamilies that predominantly signal through discrete signaling pathways. The BMP subfamily ligand DBL-1 has been well studied for its role in the innate immune response in C. elegans. Here we show that all five TGF-β ligands play a role in the immune response. We also demonstrate that multiple TGF-β ligands act cooperatively as part of this response. We show that the two BMP-like ligands - DBL-1 and TIG-2 - function independently of each other in the immune response, while TIG-2/BMP and the TGF-β/Activin-like ligand TIG-3 function cooperatively. Structural modeling supports the potential for TIG-2 and TIG-3 to form heterodimers. Finally, we show that canonical DBL-1/BMP receptor and Smad signal transducers function in the response to bacterial pathogens, while components of the DAF-7 TGF-β/Activin signaling pathway do not play a role in survival. These results demonstrate a novel potential for BMP and TGF-β/Activin subfamily ligands to interact, and may provide a mechanism for distinguishing the developmental and homeostatic functions of these ligands from an acute response such as the innate immune response to bacterial pathogens.
Figures



Similar articles
-
TGF-β ligand cross-subfamily interactions in the response of Caenorhabditis elegans to a bacterial pathogen.PLoS Genet. 2024 Jun 14;20(6):e1011324. doi: 10.1371/journal.pgen.1011324. eCollection 2024 Jun. PLoS Genet. 2024. PMID: 38875298 Free PMC article.
-
Transforming Growth Factor-β Family Ligands Can Function as Antagonists by Competing for Type II Receptor Binding.J Biol Chem. 2016 May 13;291(20):10792-804. doi: 10.1074/jbc.M115.713487. Epub 2016 Mar 9. J Biol Chem. 2016. PMID: 26961869 Free PMC article.
-
Regulation of genes affecting body size and innate immunity by the DBL-1/BMP-like pathway in Caenorhabditis elegans.BMC Dev Biol. 2010 Jun 7;10:61. doi: 10.1186/1471-213X-10-61. BMC Dev Biol. 2010. PMID: 20529267 Free PMC article.
-
Bone morphogenetic proteins.Growth Factors. 2004 Dec;22(4):233-41. doi: 10.1080/08977190412331279890. Growth Factors. 2004. PMID: 15621726 Review.
-
TGF-β signaling in C. elegans.WormBook. 2013 Jul 10:1-34. doi: 10.1895/wormbook.1.22.2. WormBook. 2013. PMID: 23908056 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
