This is a preprint.
Early Elevated IFNα Identified as the Key Mediator of HIV Pathogenesis and its low level a Hallmark of Elite Controllers
- PMID: 37215045
- PMCID: PMC10197726
- DOI: 10.21203/rs.3.rs-2813601/v1
Early Elevated IFNα Identified as the Key Mediator of HIV Pathogenesis and its low level a Hallmark of Elite Controllers
Update in
-
Early elevated IFNα is a key mediator of HIV pathogenesis.Commun Med (Lond). 2024 Mar 19;4(1):53. doi: 10.1038/s43856-024-00454-6. Commun Med (Lond). 2024. PMID: 38504106 Free PMC article.
Abstract
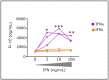
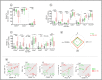
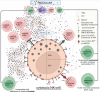
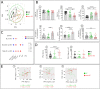
Advances in HIV therapy came from understanding its replication. Further progress toward "functional cure" -no therapy needed as found in Elite Controllers (EC)- may come from insights in pathogenesis and avoidance by EC. Here we show that all immune cells from HIV-infected persons are impaired in non-EC, but not in EC. Since HIV infects few cell types, these results suggest an additional mediator of pathogenesis. We identify that mediator as elevated pathogenic IFNα, controlled by EC likely by their preserved potent NK-cells and later by other killer cells. Since the earliest days of infection predict outcome genetic or chance events must be key to EC, and since we found no unique immune parameter at the onset, we suggest a chance infection with a lower HIV inoculum. These results offer an additional approach toward functional cure: a judicious targeting of IFNα for all non-EC patients.
Figures







Similar articles
-
Early elevated IFNα is a key mediator of HIV pathogenesis.Commun Med (Lond). 2024 Mar 19;4(1):53. doi: 10.1038/s43856-024-00454-6. Commun Med (Lond). 2024. PMID: 38504106 Free PMC article.
-
IFNα induces CCR5 in CD4+ T cells of HIV patients causing pathogenic elevation.Commun Med (Lond). 2024 Mar 19;4(1):52. doi: 10.1038/s43856-024-00453-7. Commun Med (Lond). 2024. PMID: 38504093 Free PMC article.
-
IFNα induces CCR5 in CD4+ T-cells, causing its anti- HIV inefficiency and its subsequent pathogenic elevation, partially controlled by anti-HIV therapy.Res Sq [Preprint]. 2023 May 11:rs.3.rs-2813616. doi: 10.21203/rs.3.rs-2813616/v1. Res Sq. 2023. Update in: Commun Med (Lond). 2024 Mar 19;4(1):52. doi: 10.1038/s43856-024-00453-7. PMID: 37214795 Free PMC article. Updated. Preprint.
-
Elite controllers: A heterogeneous group of HIV-infected patients.Virulence. 2020 Dec;11(1):889-897. doi: 10.1080/21505594.2020.1788887. Virulence. 2020. PMID: 32698654 Free PMC article. Review.
-
Learning to Be Elite: Lessons From HIV-1 Controllers and Animal Models on Trained Innate Immunity and Virus Suppression.Front Immunol. 2022 Apr 27;13:858383. doi: 10.3389/fimmu.2022.858383. eCollection 2022. Front Immunol. 2022. PMID: 35572502 Free PMC article. Review.
References
-
- Deeks S. G. & Walker B. D. Human Immunodeficiency Virus Controllers: Mechanisms of Durable Virus Control in the Absence of Antiretroviral Therapy. Immunity 27, 406–416 (2007). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

