The dynamical organization of the core pluripotency transcription factors responds to differentiation cues in early S-phase
- PMID: 37215075
- PMCID: PMC10192714
- DOI: 10.3389/fcell.2023.1125015
The dynamical organization of the core pluripotency transcription factors responds to differentiation cues in early S-phase
Abstract
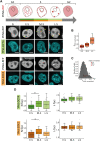
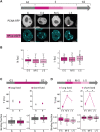
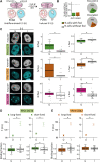
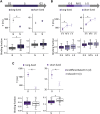
DNA replication in stem cells is a major challenge for pluripotency preservation and cell fate decisions. This process involves massive changes in the chromatin architecture and the reorganization of many transcription-related molecules in different spatial and temporal scales. Pluripotency is controlled by the master transcription factors (TFs) OCT4, SOX2 and NANOG that partition into condensates in the nucleus of embryonic stem cells. These condensates are proposed to play relevant roles in the regulation of gene expression and the maintenance of pluripotency. Here, we asked whether the dynamical distribution of the pluripotency TFs changes during the cell cycle, particularly during DNA replication. Since the S phase is considered to be a window of opportunity for cell fate decisions, we explored if differentiation cues in G1 phase trigger changes in the distribution of these TFs during the subsequent S phase. Our results show a spatial redistribution of TFs condensates during DNA replication which was not directly related to chromatin compaction. Additionally, fluorescence fluctuation spectroscopy revealed TF-specific, subtle changes in the landscape of TF-chromatin interactions, consistent with their particularities as key players of the pluripotency network. Moreover, we found that differentiation stimuli in the preceding G1 phase triggered a relatively fast and massive reorganization of pluripotency TFs in early-S phase. Particularly, OCT4 and SOX2 condensates dissolved whereas the lifetimes of TF-chromatin interactions increased suggesting that the reorganization of condensates is accompanied with a change in the landscape of TF-chromatin interactions. Notably, NANOG showed impaired interactions with chromatin in stimulated early-S cells in line with its role as naïve pluripotency TF. Together, these findings provide new insights into the regulation of the core pluripotency TFs during DNA replication of embryonic stem cells and highlight their different roles at early differentiation stages.
Keywords: DNA replication; cell cycle; condensates; differentiation; embryonic stem cells; pluripotency transcription factors.
Copyright © 2023 Oses, Francia, Verneri, Vazquez Echegaray, Guberman and Levi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Dynamical reorganization of the pluripotency transcription factors Oct4 and Sox2 during early differentiation of embryonic stem cells.Sci Rep. 2020 Mar 23;10(1):5195. doi: 10.1038/s41598-020-62235-0. Sci Rep. 2020. PMID: 32251342 Free PMC article.
-
Pluripotency transcription factors at the focus: the phase separation paradigm in stem cells.Biochem Soc Trans. 2021 Dec 17;49(6):2871-2878. doi: 10.1042/BST20210856. Biochem Soc Trans. 2021. PMID: 34812855
-
A Simple Method for Generating Light-induced Clusters of Transcription Factors: Effects on the Nuclear Distribution of OCT4 and on its Interactions With Chromatin.J Mol Biol. 2025 Jun 15;437(12):169118. doi: 10.1016/j.jmb.2025.169118. Epub 2025 Mar 31. J Mol Biol. 2025. PMID: 40174669
-
SOX2, OCT4 and NANOG: The core embryonic stem cell pluripotency regulators in oral carcinogenesis.J Oral Maxillofac Pathol. 2020 May-Aug;24(2):368-373. doi: 10.4103/jomfp.JOMFP_22_20. Epub 2020 Sep 9. J Oral Maxillofac Pathol. 2020. PMID: 33456249 Free PMC article. Review.
-
Role of Oct4 in maintaining and regaining stem cell pluripotency.Stem Cell Res Ther. 2010 Dec 14;1(5):39. doi: 10.1186/scrt39. Stem Cell Res Ther. 2010. PMID: 21156086 Free PMC article. Review.
Cited by
-
Differential cell architecture and microenvironmental responses of pretumoral and tumoral cellular models exposed to coverslip-induced hypoxia.Histochem Cell Biol. 2025 Jan 3;163(1):23. doi: 10.1007/s00418-024-02350-5. Histochem Cell Biol. 2025. PMID: 39751684
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

