Evidence for broad cross-reactivity of the SARS-CoV-2 NSP12-directed CD4+ T-cell response with pre-primed responses directed against common cold coronaviruses
- PMID: 37215095
- PMCID: PMC10196118
- DOI: 10.3389/fimmu.2023.1182504
Evidence for broad cross-reactivity of the SARS-CoV-2 NSP12-directed CD4+ T-cell response with pre-primed responses directed against common cold coronaviruses
Abstract
Introduction: The nonstructural protein 12 (NSP12) of the severe acute respiratory syndrome coronavirus type 2 (SARS-CoV-2) has a high sequence identity with common cold coronaviruses (CCC).
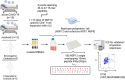
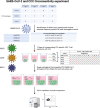
Methods: Here, we comprehensively assessed the breadth and specificity of the NSP12-specific T-cell response after in vitro T-cell expansion with 185 overlapping 15-mer peptides covering the entire SARS-CoV-2 NSP12 at single-peptide resolution in a cohort of 27 coronavirus disease 2019 (COVID-19) patients. Samples of nine uninfected seronegative individuals, as well as five pre-pandemic controls, were also examined to assess potential cross-reactivity with CCCs.
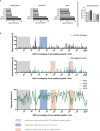
Results: Surprisingly, there was a comparable breadth of individual NSP12 peptide-specific CD4+ T-cell responses between COVID-19 patients (mean: 12.82 responses; range: 0-25) and seronegative controls including pre-pandemic samples (mean: 12.71 responses; range: 0-21). However, the NSP12-specific T-cell responses detected in acute COVID-19 patients were on average of a higher magnitude. The most frequently detected CD4+ T-cell peptide specificities in COVID-19 patients were aa236-250 (37%) and aa246-260 (44%), whereas the peptide specificities aa686-700 (50%) and aa741-755 (36%), were the most frequently detected in seronegative controls. In CCC-specific peptide-expanded T-cell cultures of seronegative individuals, the corresponding SARS-CoV-2 NSP12 peptide specificities also elicited responses in vitro. However, the NSP12 peptide-specific CD4+ T-cell response repertoire only partially overlapped in patients analyzed longitudinally before and after a SARS-CoV-2 infection.
Discussion: The results of the current study indicate the presence of pre-primed, cross-reactive CCC-specific T-cell responses targeting conserved regions of SARS-CoV-2, but they also underline the complexity of the analysis and the limited understanding of the role of the SARS-CoV-2 specific T-cell response and cross-reactivity with the CCCs.
Keywords: CD4+ T-cells; Cross-reactivities; NSP12; RNA-dependant RNA polymerase; SARS-CoV-2; common cold coronaviruses; epitope analysis; sequence identities.
Copyright © 2023 Westphal, Mader, Karsten, Cords, Knapp, Schulte, Hermanussen, Peine, Ditt, Grifoni, Addo, Huber, Sette, Lütgehetmann, Pischke, Kwok, Sidney and Schulze zur Wiesch.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Immunopeptidome profiling of human coronavirus OC43-infected cells identifies CD4 T-cell epitopes specific to seasonal coronaviruses or cross-reactive with SARS-CoV-2.PLoS Pathog. 2023 Jul 27;19(7):e1011032. doi: 10.1371/journal.ppat.1011032. eCollection 2023 Jul. PLoS Pathog. 2023. PMID: 37498934 Free PMC article.
-
SARS-CoV-2-Seronegative Subjects Target CTL Epitopes in the SARS-CoV-2 Nucleoprotein Cross-Reactive to Common Cold Coronaviruses.Front Immunol. 2021 Apr 28;12:627568. doi: 10.3389/fimmu.2021.627568. eCollection 2021. Front Immunol. 2021. PMID: 33995351 Free PMC article. Clinical Trial.
-
Differential T-Cell Reactivity to Endemic Coronaviruses and SARS-CoV-2 in Community and Health Care Workers.J Infect Dis. 2021 Jul 2;224(1):70-80. doi: 10.1093/infdis/jiab176. J Infect Dis. 2021. PMID: 33822097 Free PMC article.
-
SARS-CoV-2-Specific T Cell Responses in Patients with COVID-19 and Unexposed Individuals.Immune Netw. 2021 Feb 17;21(1):e2. doi: 10.4110/in.2021.21.e2. eCollection 2021 Feb. Immune Netw. 2021. PMID: 33728095 Free PMC article. Review.
-
Architecture of the SARS-CoV-2-specific T cell repertoire.Front Immunol. 2023 Mar 20;14:1070077. doi: 10.3389/fimmu.2023.1070077. eCollection 2023. Front Immunol. 2023. PMID: 37020560 Free PMC article. Review.
Cited by
-
Heterotypic responses against nsp12/nsp13 from prior SARS-CoV-2 infection associates with lower subsequent endemic coronavirus incidence.bioRxiv [Preprint]. 2023 Oct 24:2023.10.23.563621. doi: 10.1101/2023.10.23.563621. bioRxiv. 2023. Update in: Sci Transl Med. 2024 Jun 12;16(751):eado7588. doi: 10.1126/scitranslmed.ado7588. PMID: 37961343 Free PMC article. Updated. Preprint.
-
Heterotypic immunity from prior SARS-CoV-2 infection but not COVID-19 vaccination associates with lower endemic coronavirus incidence.Sci Transl Med. 2024 Jun 12;16(751):eado7588. doi: 10.1126/scitranslmed.ado7588. Epub 2024 Jun 12. Sci Transl Med. 2024. PMID: 38865483 Free PMC article.
-
Human coronavirus OC43-elicited CD4+ T cells protect against SARS-CoV-2 in HLA transgenic mice.Nat Commun. 2024 Jan 26;15(1):787. doi: 10.1038/s41467-024-45043-2. Nat Commun. 2024. PMID: 38278784 Free PMC article.
-
Detection of Antibodies against Endemic and SARS-CoV-2 Coronaviruses with Short Peptide Epitopes.Vaccines (Basel). 2023 Aug 23;11(9):1403. doi: 10.3390/vaccines11091403. Vaccines (Basel). 2023. PMID: 37766081 Free PMC article.
-
Development and Validation of a Highly Sensitive Multiplex Immunoassay for SARS-CoV-2 Humoral Response Monitorization: A Study of the Antibody Response in COVID-19 Patients with Different Clinical Profiles during the First and Second Waves in Cadiz, Spain.Microorganisms. 2023 Dec 16;11(12):2997. doi: 10.3390/microorganisms11122997. Microorganisms. 2023. PMID: 38138141 Free PMC article.
References
-
- Heide J, Schulte S, Kohsar M, Brehm TT, Herrmann M, Karsten H, et al. Broadly directed SARS-CoV-2-specific CD4+ T cell response includes frequently detected peptide specificities within the membrane and nucleoprotein in patients with acute and resolved COVID-19. PLoS Pathog (2021) 17(9):e1009842. doi: 10.1371/journal.ppat.1009842 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

