Identification of hub cuproptosis related genes and immune cell infiltration characteristics in periodontitis
- PMID: 37215133
- PMCID: PMC10196202
- DOI: 10.3389/fimmu.2023.1164667
Identification of hub cuproptosis related genes and immune cell infiltration characteristics in periodontitis
Erratum in
-
Corrigendum: Identification of hub cuproptosis related genes and immune cell infiltration characteristics in periodontitis.Front Immunol. 2023 Oct 3;14:1281051. doi: 10.3389/fimmu.2023.1281051. eCollection 2023. Front Immunol. 2023. PMID: 37854598 Free PMC article.
Abstract
Introduction: Periodontitis is an inflammatory disease and its molecular mechanisms is not clear. A recently discovered cell death pathway called cuproptosis, may related to the disease.
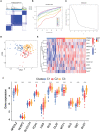
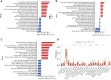
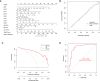
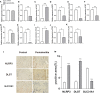
Methods: The datasets GSE10334 of human periodontitis and control were retrieved from the Gene Expression Omnibus database (GEO) for analysis.Following the use of two machine learning algorithms, least absolute shrinkage and selection operator (LASSO) and support vector machine-recursive feature removal (SVM-RFE) were used to find CRG-based signature. Then the Receiver operating characteristic (ROC) curves was used to evaluate the gene signature's discriminatory ability. The CIBERSORT deconvolution algorithm was used to study the link between hub genes and distinct types of immune cells. Next, the association of the CRGs with immune cells in periodontitis and relevant clusters of cuproptosis were found. The link between various clusters was ascertained by the GSVA and CIBERSORT deconvolution algorithm. Finally, An external dataset (GSE16134) was used to confirm the diagnosis capacity of the identified biomarkers. In addition, clinical samples were examined using qRT-PCR and immunohistochemistry to verifiy the expression of genes related to cuprotosis in periodontitis and the signature may better predict the periodontitis.
Results: 15 periodontitis-related DE-CRGs were found,then 11-CRG-based signature was found by using of LASSO and SVM-RFE. ROC curves also supported the value of signature. CIBERSORT results of immune cell signature in periodontitis showed that signature genes is a crucial component of the immune response.The relevant clusters of cuproptosis found that the NFE2L2, SLC31A1, FDX1,LIAS, DLD, DLAT, and DBT showed a highest expression levels in Cluster2 ,while the NLRP3, MTF1, and DLST displayed the lowest level in Cluster 2 but the highest level in Cluster1. The GSVA results also showed that the 11 cuproptosis diagnostic gene may regulate the periodontitis by affecting immune cells. The external dataset (GSE16134) confirm the diagnosis capacity of the identified biomarkers, and clinical samples examined by qRT-PCR and immunohistochemistry also verified that these cuprotosis related signiture genes in periodontitis may better predict the periodontitis.
Conclusion: These findings have important implications for the cuproptosis and periodontitis, and highlight further research is needed to better understand the mechanisms underlying this relationship between the cuproptosis and periodontitis.
Keywords: cuproptosis; hub; immune; infiltration; periodontitis.
Copyright © 2023 Liu, Ge, Chu, Cai, Wu, Gong and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

