Chromosomal microarray analysis supplements exome sequencing to diagnose children with suspected inborn errors of immunity
- PMID: 37215141
- PMCID: PMC10196392
- DOI: 10.3389/fimmu.2023.1172004
Chromosomal microarray analysis supplements exome sequencing to diagnose children with suspected inborn errors of immunity
Abstract
Purpose: Though copy number variants (CNVs) have been suggested to play a significant role in inborn errors of immunity (IEI), the precise nature of this role remains largely unexplored. We sought to determine the diagnostic contribution of CNVs using genome-wide chromosomal microarray analysis (CMA) in children with IEI.
Methods: We performed exome sequencing (ES) and CMA for 332 unrelated pediatric probands referred for evaluation of IEI. The analysis included primary, secondary, and incidental findings.
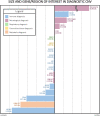
Results: Of the 332 probands, 134 (40.4%) received molecular diagnoses. Of these, 116/134 (86.6%) were diagnosed by ES alone. An additional 15/134 (11.2%) were diagnosed by CMA alone, including two likely de novo changes. Three (2.2%) participants had diagnostic molecular findings from both ES and CMA, including two compound heterozygotes and one participant with two distinct diagnoses. Half of the participants with CMA contribution to diagnosis had CNVs in at least one non-immune gene, highlighting the clinical complexity of these cases. Overall, CMA contributed to 18/134 diagnoses (13.4%), increasing the overall diagnostic yield by 15.5% beyond ES alone.
Conclusion: Pairing ES and CMA can provide a comprehensive evaluation to clarify the complex factors that contribute to both immune and non-immune phenotypes. Such a combined approach to genetic testing helps untangle complex phenotypes, not only by clarifying the differential diagnosis, but in some cases by identifying multiple diagnoses contributing to the overall clinical presentation.
Trial registration: ClinicalTrials.gov NCT03206099.
Keywords: copy number; diagnosis; genetic; immunity; microarray; pediatric; primary immunodeficiency; sequencing.
Copyright © 2023 Beers, Similuk, Ghosh, Seifert, Jamal, Kamen, Setzer, Jodarski, Duncan, Hunt, Mixer, Cao, Bi, Veltri, Karlins, Zhang, Li, Oler, Jevtich, Yu, Hullfish, Bielekova, Frischmeyer-Guerrerio, Dang Do, Huryn, Olivier, Su, Lyons, Zerbe, Rao, Keller, Freeman, Holland, Franco, Walkiewicz and Yan.
Conflict of interest statement
WB is an employee of Baylor Genetics. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer IC declared a shared affiliation, with no collaboration, with one of the authors, WB, to the handling editor at the time of the review.
Figures

References
-
- Tangye SG, Al-Herz W, Bousfiha A, Chatila T, Cunningham-Rundles C, Etzioni A, et al. . Human inborn errors of immunity: 2019 update on the classification from the international union of immunological societies expert committee. J Clin Immunol (2020) 40(1):24–64. doi: 10.1007/s10875-019-00737-x - DOI - PMC - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

