Strategies for precise gene edits in mammalian cells
- PMID: 37215153
- PMCID: PMC10192336
- DOI: 10.1016/j.omtn.2023.04.012
Strategies for precise gene edits in mammalian cells
Abstract
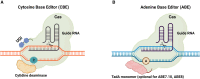
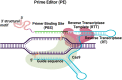
CRISPR-Cas technologies have the potential to revolutionize genetic medicine. However, work is still needed to make this technology clinically efficient for gene correction. A barrier to making precise genetic edits in the human genome is controlling how CRISPR-Cas-induced DNA breaks are repaired by the cell. Since error-prone non-homologous end-joining is often the preferred cellular repair pathway, CRISPR-Cas-induced breaks often result in gene disruption. Homology-directed repair (HDR) makes precise genetic changes and is the clinically desired pathway, but this repair pathway requires a homology donor template and cycling cells. Newer editing strategies, such as base and prime editing, can affect precise repair for relatively small edits without requiring HDR and circumvent cell cycle dependence. However, these technologies have limitations in the extent of genetic editing and require the delivery of bulky cargo. Here, we discuss the pros and cons of precise gene correction using CRISPR-Cas-induced HDR, as well as base and prime editing for repairing small mutations. Finally, we consider emerging new technologies, such as recombination and transposases, which can circumvent both cell cycle and cellular DNA repair dependence for editing the genome.
Keywords: CRISPR-Cas9; MT: RNA/DNA Editing; base editing; gene editing; homology-directed repair; prime editing.
© 2023 The Author(s).
Conflict of interest statement
P.M. has patents and royalties with CSL Behring and Aruvant Sciences.
Figures


References
-
- Chapman J.R., Taylor M.R.G., Boulton S.J. Playing the end game: DNA double-strand break repair pathway choice. Mol. Cell. 2012;47:497–510. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

